6FWZ
 
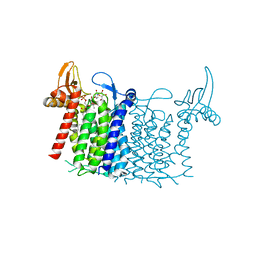 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with UDP-GlcNAc | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, MAGNESIUM ION, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-07 | | Release date: | 2018-07-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
6MC1
 
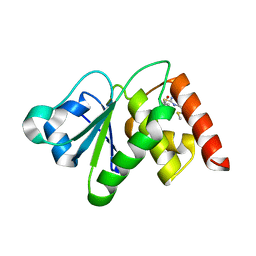 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
8GH5
 
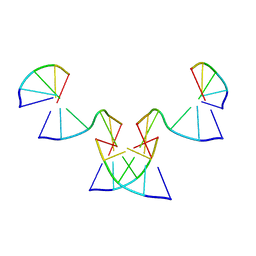 | | Implementing Logic Gates in DNA Crystal Engineering | | Descriptor: | DNA (5'-D(*AP*GP*AP*CP*G)-3'), DNA (5'-D(*CP*TP*AP*CP*G)-3'), DNA (5'-D(P*CP*GP*TP*GP*GP*A)-3'), ... | | Authors: | Zhang, C, Paluzzi, V.E, Sha, R, Jonoska, N, Mao, C. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Implementing Logic Gates by DNA Crystal Engineering.
Adv Mater, 35, 2023
|
|
7NHF
 
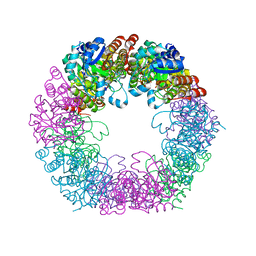 | | Crystal structure of Arabidopsis thaliana Pdx1K166R | | Descriptor: | PHOSPHATE ION, Pyridoxal 5'-phosphate synthase subunit PDX1.3 | | Authors: | Rodrigues, M.J, Zhang, Y, Bolton, R, Evans, G, Giri, N, Royant, A, Begley, T, Ealick, S.E, Tews, I. | | Deposit date: | 2021-02-10 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Trapping and structural characterisation of a covalent intermediate in vitamin B 6 biosynthesis catalysed by the Pdx1 PLP synthase.
Rsc Chem Biol, 3, 2022
|
|
6XNP
 
 | | Crystal Structure of Human STING CTD complex with SR-717 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-difluoro-2-{[6-(1H-imidazol-1-yl)pyridazine-3-carbonyl]amino}benzoic acid, GLYCEROL, ... | | Authors: | Chin, E.N, Yu, C, Wolan, D.W, Petrassi, H.M, Lairson, L.L. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Antitumor activity of a systemic STING-activating non-nucleotide cGAMP mimetic.
Science, 369, 2020
|
|
6P1L
 
 | | Crystal structure of EGFR in complex with EAI045 | | Descriptor: | (2R)-2-(5-fluoro-2-hydroxyphenyl)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-05-20 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery and Optimization of Dibenzodiazepinones as Allosteric Mutant-Selective EGFR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7O8F
 
 | | NmHR dark state structure determined by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8H
 
 | | NmHR light state structure at 10 ns after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8G
 
 | | NmHR light state structure at 10 ps after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7NWW
 
 | | CspA-27 cotranslational folding intermediate 1 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Samatova, E, Macher, M, Liutkute, M, Gil-Carton, D, Novacek, J, Valle, M, Rodnina, M.V. | | Deposit date: | 2021-03-17 | | Release date: | 2022-01-19 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | A switch from alpha-helical to beta-strand conformation during co-translational protein folding.
Embo J., 41, 2022
|
|
7A9Z
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 1 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6XP0
 
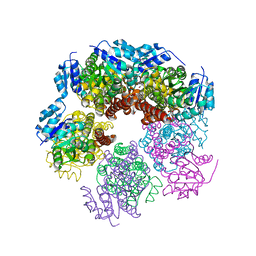 | |
6TL4
 
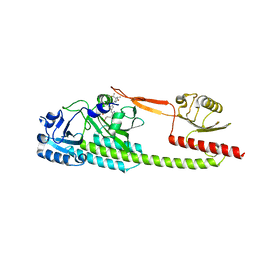 | | Photosensory module (PAS-GAF-PHY) of Glycine max phyB | | Descriptor: | PHYCOCYANOBILIN, Phytochrome | | Authors: | Nagano, S, Guan, K, Shenkutie, S.M, Hughes, J.E. | | Deposit date: | 2019-12-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into photoactivation and signalling in plant phytochromes.
Nat.Plants, 6, 2020
|
|
6FRN
 
 | | Structure of F420H2 oxidase (FprA) co-crystallized with 10mM Tb-Xo4 and calcium chloride | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CALCIUM ION, F420H2 oxidase (FprA), ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Wagner, T, Shima, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2018-02-16 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
7A9Y
 
 | | Structural comparison of cellular retinoic acid binding protein I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | Cellular retinoic acid-binding protein 1, GLYCEROL, MYRISTIC ACID, ... | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
8UZU
 
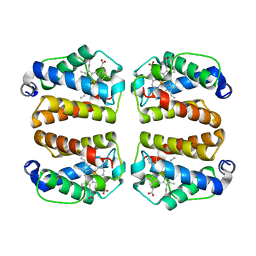 | | Crystal structure of Shewanella benthica Group 1 truncated hemoglobin L80A C51S C71S variant | | Descriptor: | CYANIDE ION, Group 1 truncated hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schultz, T.D, Martinez, J.E, Siegler, M.A, Schlessman, J.L, Lecomte, J.T.J. | | Deposit date: | 2023-11-16 | | Release date: | 2024-04-03 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Extremophilic hemoglobins: The structure of Shewanella benthica truncated hemoglobin N.
J.Biol.Chem., 301, 2025
|
|
7NTQ
 
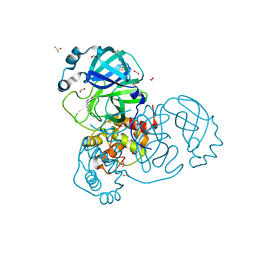 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
7Q1B
 
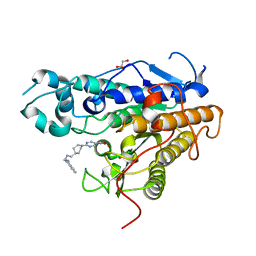 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, GLYCEROL, Histone deacetylase DAC2, ... | | Authors: | Marek, M, Ramos-Morales, E, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
7NTW
 
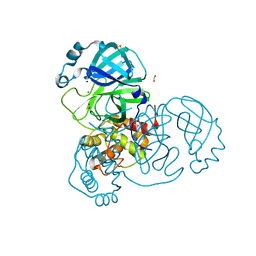 | |
7NTT
 
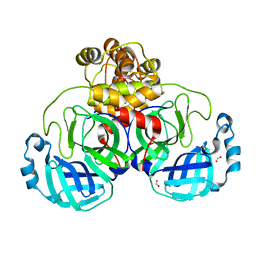 | |
6PKA
 
 | |
6PFQ
 
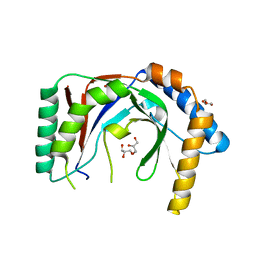 | |
8BYA
 
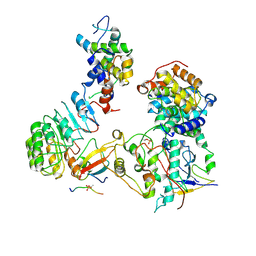 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
6ZT5
 
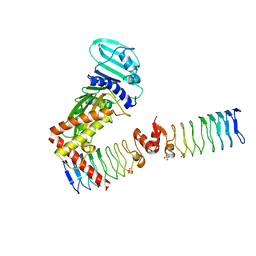 | | Complex between a homodimer of Mycobacterium smegmatis MfpA and a single copy of the N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit | | Descriptor: | DNA gyrase subunit B, Pentapeptide repeat protein MfpA, SULFATE ION | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZT3
 
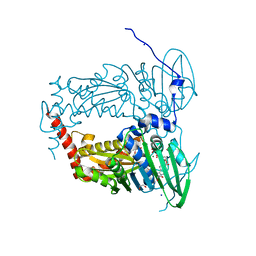 | | N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit complexed with ADPNP | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, MAGNESIUM ION, ... | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
