7VGV
 
 | | Anion free form of light-driven chloride ion-pumping rhodopsin, NM-R3, structure determined by serial femtosecond crystallography at SACLA | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, HEXADECANE, ... | | Authors: | Hosaka, T, Nango, E, Nakane, T, Luo, F, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-09-18 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4O4K
 
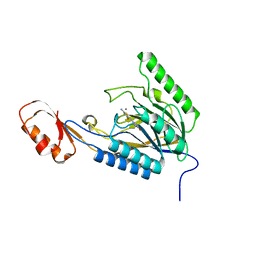 | | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities | | Descriptor: | (5~{E})-2-azanylidene-5-[(4-hydroxyphenyl)methylidene]-1,3-thiazolidin-4-one, Exonuclease, putative, ... | | Authors: | Shibata, A, Moiani, D, Arvai, A.S, Perry, J, Harding, S.M, Genois, M, Maity, R, Rossum-Fikkert, S, Kertokalio, A, Romoli, F, Ismail, A, Ismalaj, E, Petricci, E, Neale, M.J, Bristow, R.G, Masson, J, Wyman, C, Jeggo, P.A, Tainer, J.A. | | Deposit date: | 2013-12-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | DNA Double-Strand Break Repair Pathway Choice Is Directed by Distinct MRE11 Nuclease Activities.
Mol.Cell, 53, 2014
|
|
8S9T
 
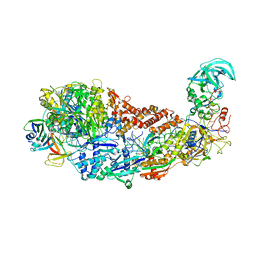 | | CRISPR-Cas type III-D effector complex | | Descriptor: | CRISPR RNA, Cas10, Cas7-2x, ... | | Authors: | Schwartz, E.A, Taylor, D.W. | | Deposit date: | 2023-03-30 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | RNA targeting and cleavage by the type III-Dv CRISPR effector complex.
Nat Commun, 15, 2024
|
|
8S9U
 
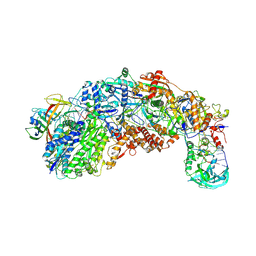 | |
4FAL
 
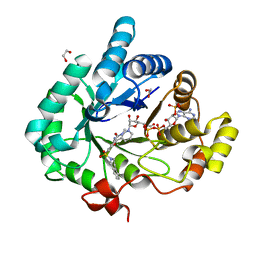 | | Crystal structure of human 17beta-hydroxysteroid dehydrogenase type 5 in complex with 3-((3,4-dihydroisoquinolin-2(1H)-yl)sulfonyl)-N-methylbenzamide (80) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)-N-methylbenzamide, Aldo-keto reductase family 1 member C3, ... | | Authors: | Turnbull, A.P, Jamieson, S.M.F, Brooke, D.G, Heinrich, D, Atwell, G.J, Silva, S, Hamilton, E.J, Rigoreau, L.J.M, Trivier, E, Soudy, C, Samlal, S.S, Owen, P.J, Schroeder, E, Raynham, T, Flanagan, J.U, Denny, W.A. | | Deposit date: | 2012-05-22 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(3,4-Dihydroisoquinolin-2(1H)-ylsulfonyl)benzoic acids; a New Class of Highly Potent and Selective Inhibitors of the Type 5 17-beta-hydroxysteroid Dehydrogenase AKR1C3
J.Med.Chem., 55, 2012
|
|
2GBH
 
 | | NMR structure of stem region of helix-35 of 23S E.coli ribosomal RNA (residues 736-760) | | Descriptor: | 5'-R(*(GMP)P*GP*GP*CP*UP*AP*AP*UP*GP*(PSU)P*UP*GP*AP*AP*AP*AP*AP*UP*UP*AP*GP*CP*CP*C)-3' | | Authors: | Bax, A, Boisbouvier, J, Bryce, D, Grishaev, A, Jaroniec, C, Miclet, E, Nikonovicz, E, O'Neil-Cabello, E, Ying, J. | | Deposit date: | 2006-03-10 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Measurement of five dipolar couplings from a single 3D NMR multiplet applied to the study of RNA dynamics.
J.Am.Chem.Soc., 126, 2004
|
|
4RL6
 
 | | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae. Northeast Structural Genomics Consortium Target SpR105 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | Authors: | Vorobiev, S, Neely, H.M, Odukwe, C.D, Seetharaman, J, Mao, L, Xiao, R, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-05 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structure of the Q04L03_STRP2 protein from Streptococcus pneumoniae.
To be Published
|
|
8SL9
 
 | | Crystal structure of Francisella tularensis HPPK-DHPS in complex with HPPK inhibitor HP-73 | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, 5'-S-[(2R,4R)-1-{2-[(2-amino-7,7-dimethyl-4-oxo-3,4,7,8-tetrahydropteridine-6-carbonyl)amino]ethyl}-2-carboxypiperidin-4-yl]-5'-thioadenosine, ... | | Authors: | Shaw, G.X, Shi, G, Cherry, S, Needle, D, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of Francisella tularensis HPPK-DHPS in complex with HPPK inhibitor HP-73
To be published
|
|
4A71
 
 | | cytochrome c peroxidase in complex with phenol | | Descriptor: | CYTOCHROME C PEROXIDASE, MITOCHONDRIAL, PHENOL, ... | | Authors: | Murphy, E.J, Metcalfe, C.L, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2011-11-10 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal Structure of Guaiacol and Phenol Bound to a Heme Peroxidase.
FEBS J., 279, 2012
|
|
8WDV
 
 | | Photosynthetic LH1-RC complex from the purple sulfur bacterium Allochromatium vinosum purified by Ca2+-DEAE | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, Antenna complex alpha/beta subunit, ... | | Authors: | Tani, K, Kanno, R, Harada, A, Kobayashi, A, Minamino, A, Nakamura, N, Ji, X.-C, Purba, E.R, Hall, M, Yu, L.-J, Madigan, M.T, Mizoguchi, A, Iwasaki, K, Humbel, B.M, Kimura, Y, Wang-Otomo, Z.-Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | High-resolution structure and biochemical properties of the LH1-RC photocomplex from the model purple sulfur bacterium, Allochromatium vinosum.
Commun Biol, 7, 2024
|
|
2GHP
 
 | | Crystal structure of the N-terminal 3 RNA binding domains of the yeast splicing factor Prp24 | | Descriptor: | U4/U6 snRNA-associated splicing factor PRP24 | | Authors: | Bae, E, Wesenberg, G.E, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and interactions of the first three RNA recognition motifs of splicing factor prp24.
J.Mol.Biol., 367, 2007
|
|
1NNP
 
 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | Authors: | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
3HV0
 
 | |
6FUJ
 
 | | Complement factor D in complex with the inhibitor N-(3'-(aminomethyl)-[1,1'-biphenyl]-3-yl)-3-methylbutanamide | | Descriptor: | Complement factor D, ~{N}-[3-[3-(aminomethyl)phenyl]phenyl]-3-methyl-butanamide | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
4RXV
 
 | | The crystal structure of the N-terminal fragment of uncharacterized protein from Legionella pneumophila | | Descriptor: | hypothetical protein lpg0944 | | Authors: | Nocek, B, Cuff, M, Evdokimova, E, Egorova, O, Joachimiak, A, Ensminger, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-12-12 | | Release date: | 2015-04-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Diverse mechanisms of metaeffector activity in an intracellular bacterial pathogen, Legionella pneumophila.
Mol Syst Biol, 12, 2016
|
|
6FVX
 
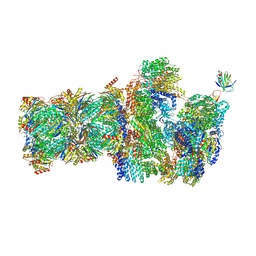 | | 26S proteasome, s5 state | | Descriptor: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | Authors: | Eisele, M.R, Reed, R.G, Rudack, T, Schweitzer, A, Beck, F, Nagy, I, Pfeifer, G, Plitzko, J.M, Baumeister, W, Tomko, R.J, Sakata, E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating.
Cell Rep, 24, 2018
|
|
7JSW
 
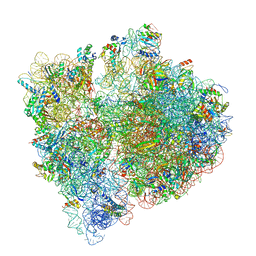 | |
8PUM
 
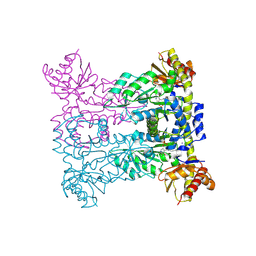 | |
4TPG
 
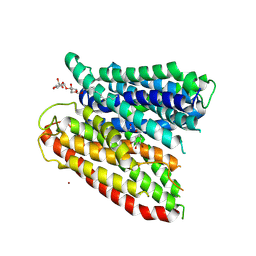 | | Selectivity mechanism of a bacterial homologue of the human drug peptide transporters PepT1 and PepT2 | | Descriptor: | Ala-L-3-Br-Tyr-Ala, DODECYL-BETA-D-MALTOSIDE, Proton:oligopeptide symporter POT family, ... | | Authors: | Guettou, F, Quistgaard, E.M, Raba, M, Moberg, P, Low, C, Nordlund, P. | | Deposit date: | 2014-06-07 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Selectivity mechanism of a bacterial homolog of the human drug-peptide transporters PepT1 and PepT2.
Nat.Struct.Mol.Biol., 21, 2014
|
|
7VUM
 
 | | Crystal structure of SSB complexed with que | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, Single-stranded DNA-binding protein | | Authors: | Lin, E.S, Huang, Y.H, Huang, C.Y. | | Deposit date: | 2021-11-03 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | A Complexed Crystal Structure of a Single-Stranded DNA-Binding Protein with Quercetin and the Structural Basis of Flavonol Inhibition Specificity.
Int J Mol Sci, 23, 2022
|
|
7BOH
 
 | | Complete Bacterial 30S ribosomal subunit assembly complex state E (+RbfA)(Consensus Refinement) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
2BKT
 
 | | crystal structure of renin-pf00257567 complex | | Descriptor: | 1-{4-[3-(2-METHOXY-BENZYLOXY)-PROPOXY]-PHENYL}-6-(1,2,,3,4-TETRAHYDRO-QUINOLIN-7-YLOXYMETHYL)-PIPERAZIN-2-ONE, RENIN | | Authors: | Powell, N.A, Clay, E.H, Holsworth, D.D, Edmunds, J.J, Bryant, J.W, Ryan, J.M, Jalaie, M, Zhang, E. | | Deposit date: | 2005-02-18 | | Release date: | 2006-04-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Equipotent Activity in Both Enantiomers of a Series of Ketopiperazine-Based Renin Inhibitors
Bioorg.Med.Chem.Lett., 15, 2005
|
|
4S3M
 
 | |
5CRO
 
 | |
1NMO
 
 | | Structural genomics, protein ybgI, unknown function | | Descriptor: | FE (III) ION, Hypothetical protein ybgI | | Authors: | Ladner, J.E, Obmolova, G, Teplyakov, A, Khil, P.P, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-10 | | Release date: | 2004-01-20 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Escherichia coli Protein ybgI, a toroidal structure with a dinuclear metal site
BMC Struct.Biol., 3, 2003
|
|
