7YYP
 
 | | Structure of aIF5B from Pyrococcus abyssi complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, NITRATE ION, Probable translation initiation factor IF-2 | | Authors: | Bourgeois, G, Schmitt, E, Mechulam, Y, Coureux, P.D, Kazan, R. | | Deposit date: | 2022-02-18 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
5DN6
 
 | | ATP synthase from Paracoccus denitrificans | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F0 subcomplex C subunit, ... | | Authors: | Morales-Rios, E, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-09-09 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structure of ATP synthase from Paracoccus denitrificans determined by X-ray crystallography at 4.0 angstrom resolution.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7NN8
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1H-indole-3-carbonitrile. | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indole-3-carbonitrile, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6F5U
 
 | | CRYSTAL STRUCTURE OF EBOLAVIRUS GLYCOPROTEIN IN COMPLEX WITH BEPRIDIL | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bepridil, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Target Identification and Mode of Action of Four Chemically Divergent Drugs against Ebolavirus Infection.
J. Med. Chem., 61, 2018
|
|
7NN7
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with dimethyl 5-hydroxyisophthalate. | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, SULFATE ION, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.172 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6CFJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with histidyl-CAM and bound to mRNA and A-, P-, and E-site tRNAs at 2.8A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Tereshchenkov, A.G, Dobosz-Bartoszek, M, Osterman, I.A, Marks, J, Sergeeva, V.A, Kasatsky, P, Komarova, E.S, Stavrianidi, A.N, Rodin, I.A, Konevega, A.L, Sergiev, P.V, Sumbatyan, N.V, Mankin, A.S, Bogdanov, A.A, Polikanov, Y.S. | | Deposit date: | 2018-02-15 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding and Action of Amino Acid Analogs of Chloramphenicol upon the Bacterial Ribosome.
J. Mol. Biol., 430, 2018
|
|
1VIF
 
 | | STRUCTURE OF DIHYDROFOLATE REDUCTASE | | Descriptor: | DIHYDROFOLATE REDUCTASE, FOLIC ACID | | Authors: | Narayana, N, Matthews, D.A, Howell, E.E, Xuong, N.-H. | | Deposit date: | 1996-10-03 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A plasmid-encoded dihydrofolate reductase from trimethoprim-resistant bacteria has a novel D2-symmetric active site.
Nat.Struct.Biol., 2, 1995
|
|
5KAB
 
 | | Protein Tyrosine Phosphatase 1B Delta helix 7, P185G mutant in complex with TCS401, open state | | Descriptor: | 2-(OXALYL-AMINO)-4,5,6,7-TETRAHYDRO-THIENO[2,3-C]PYRIDINE-3-CARBOXYLIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Choy, M.S, Machado, L.E.S.F, Peti, W, Page, R. | | Deposit date: | 2016-06-01 | | Release date: | 2017-03-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Conformational Rigidity and Protein Dynamics at Distinct Timescales Regulate PTP1B Activity and Allostery.
Mol. Cell, 65, 2017
|
|
5K33
 
 | | Crystal structure of extracellular domain of HER2 in complex with Fcab STAB19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Ig gamma-1 chain C region, ... | | Authors: | Humm, A, Lobner, E, Goritzer, K, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-05-19 | | Release date: | 2017-04-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
6F9Q
 
 | | Binary complex of a 7S-cis-cis-nepetalactol cyclase from Nepeta mussinii with NAD+ | | Descriptor: | 7S-cis-cis-nepetalactol cyclase, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Lichman, B.R, Kamileen, M.O, Titchiner, G, Saalbach, G, Stevenson, C.E.M, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Uncoupled activation and cyclization in catmint reductive terpenoid biosynthesis.
Nat. Chem. Biol., 15, 2019
|
|
6VY4
 
 | |
5ED7
 
 | | Crystal Structure of HSV-1 UL21 C-terminal Domain | | Descriptor: | CHLORIDE ION, Tegument protein UL21 | | Authors: | Metrick, C.M, Heldwein, E.E. | | Deposit date: | 2015-10-20 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Novel Structure and Unexpected RNA-Binding Ability of the C-Terminal Domain of Herpes Simplex Virus 1 Tegument Protein UL21.
J.Virol., 90, 2016
|
|
6LMI
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-chromen-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase catalytic, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-25 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
9B0L
 
 | | Cryo-EM structure of Acanthamoeba polyphaga mimivirus Fanzor2 ternary complex | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*GP*GP*TP*GP*GP*CP*TP*GP*CP*CP*CP*CP*GP*AP*CP*CP*GP*TP*C)-3'), DNA (5'-D(P*CP*CP*CP*A)-3'), DNA (5'-D(P*GP*AP*CP*GP*GP*TP*CP*GP*GP*GP*C)-3'), ... | | Authors: | Qayyum, M.Z, Schargel, R.D, Tanwar, A.S, Kellogg, E.H, Kalathur, R.C. | | Deposit date: | 2024-03-12 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure of Fanzor2 reveals insights into the evolution of the TnpB superfamily.
Nat.Struct.Mol.Biol., 2024
|
|
7Z6X
 
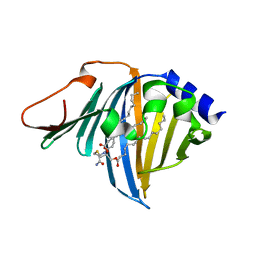 | | Complex of E. coli LolA R43L mutant and lipoprotein | | Descriptor: | Outer-membrane lipoprotein carrier protein, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Kaplan, E, Greene, N.P, Koronakis, V. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis of lipoprotein recognition by the bacterial Lol trafficking chaperone LolA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z6W
 
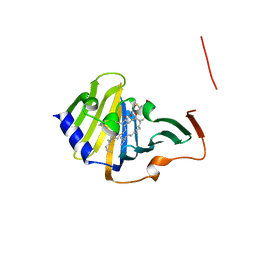 | | Complex of E. coli LolA and lipoprotein | | Descriptor: | Outer-membrane lipoprotein carrier protein, [(2~{S})-3-[(2~{S})-3-azanyl-2-(hexadecanoylamino)-3-oxidanylidene-propyl]sulfanyl-2-hexadecanoyloxy-propyl] hexadecanoate | | Authors: | Kaplan, E, Greene, N.P, Koronakis, V. | | Deposit date: | 2022-03-14 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural basis of lipoprotein recognition by the bacterial Lol trafficking chaperone LolA.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7KFN
 
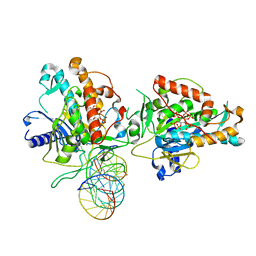 | | Structure of Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA containing a 2'-deoxy Benner's Base Z opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, Gli1 1W5 23mer RNA, Gli1 8AZ 23mer RNA, ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
J.Am.Chem.Soc., 143, 2021
|
|
6F2M
 
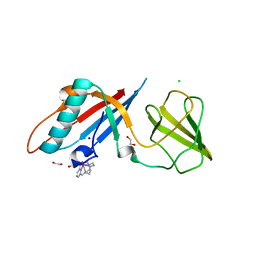 | | Structure of the bacteriophage T5 distal tail protein pb9 co-crystallized with 10mM Tb-Xo4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Distal tail protein, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Breyton, C, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
6ESQ
 
 | | Structure of the acetoacetyl-CoA thiolase/HMG-CoA synthase complex from Methanothermococcus thermolithotrophicus soaked with acetyl-CoA | | Descriptor: | CHLORIDE ION, COENZYME A, HydroxyMethylGlutaryl-CoA synthase, ... | | Authors: | Voegeli, B, Engilberge, S, Girard, E, Riobe, F, Maury, O, Erb, J.T, Shima, S, Wagner, T. | | Deposit date: | 2017-10-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
9BYJ
 
 | | Crystal Structure of Hck in complex with the Src-family kinase inhibitor A-419259 | | Descriptor: | 1,2-ETHANEDIOL, 7-[trans-4-(4-methylpiperazin-1-yl)cyclohexyl]-5-(4-phenoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DIMETHYL SULFOXIDE, ... | | Authors: | Selzer, A.M, Alvarado, J.J, Smithgall, T.E. | | Deposit date: | 2024-05-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cocrystallization of the Src-Family Kinase Hck with the ATP-Site Inhibitor A-419259 Stabilizes an Extended Activation Loop Conformation.
Biochemistry, 63, 2024
|
|
6NKE
 
 | | Wild-type GGGPS from Thermoplasma volcanium | | Descriptor: | GLYCEROL, Geranylgeranylglyceryl phosphate synthase, IMIDAZOLE, ... | | Authors: | Blank, P.N, Alderfer, K.E, Gillott, B.N, Christianson, D.W, Himmelberger, J.A. | | Deposit date: | 2019-01-07 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural studies of geranylgeranylglyceryl phosphate synthase, a prenyltransferase found in thermophilic Euryarchaeota.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6CUZ
 
 | | Engineered TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3R)-ethylserine bound as the amino-acrylate | | Descriptor: | (2E)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-2-enoic acid, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Scheele, R.A, Buller, A.R, Boville, C.E, Arnold, F.H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5K3F
 
 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
6CVN
 
 | |
5JPJ
 
 | | Crystal structure of 6aJL2-R24G | | Descriptor: | 6aJL2 protein | | Authors: | Pelaez-Aguilar, A.E, Diaz-Vilchis, A, Amero, C, Becerril, B, Rudino-Pinera, E. | | Deposit date: | 2016-05-03 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 6aJL2-R24G light chain variable domain: Does crystal packing explain amyloid fibril formation?
Biochem Biophys Rep, 20, 2019
|
|
