5MF2
 
 | | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 | | Descriptor: | CHLORIDE ION, Distal tail protein, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Lassalle, L, Arnaud, C.-A, Breyton, C, Madern, D, Coquelle, N, Maury, O, Girard, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence.
Chem Sci, 8, 2017
|
|
7UKV
 
 | | Wild type EGFR in complex with Lazertinib (YH25448) | | Descriptor: | Epidermal growth factor receptor, N-[5-{[(4P)-4-{4-[(dimethylamino)methyl]-3-phenyl-1H-pyrazol-1-yl}pyrimidin-2-yl]amino}-4-methoxy-2-(morpholin-4-yl)phenyl]propanamide | | Authors: | Beyett, T.S, Pham, C, Eck, M.J, Heppner, D.E. | | Deposit date: | 2022-04-02 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Inhibition of Mutant EGFR with Lazertinib (YH25448).
Acs Med.Chem.Lett., 13, 2022
|
|
5ESR
 
 | | Crystal structure of haloalkane dehalogenase (DccA) from Caulobacter crescentus | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Haloalkane dehalogenase, ... | | Authors: | Malashkevich, V.N, Toro, R, Mundorff, E.C, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.476 Å) | | Cite: | Biochemical characterization of two haloalkane dehalogenases: DccA from Caulobacter crescentus and DsaA from Saccharomonospora azurea.
Protein Sci., 25, 2016
|
|
1PW7
 
 | | Crystal Structure of E. coli purine nucleoside phosphorylase complexed with 9-beta-D-arabinofuranosyladenine and sulfate/phosphate | | Descriptor: | 2-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | Authors: | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | Deposit date: | 2003-06-30 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
7QP9
 
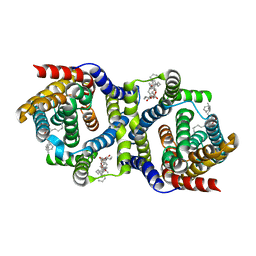 | | Outward-facing apo-form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
3I7F
 
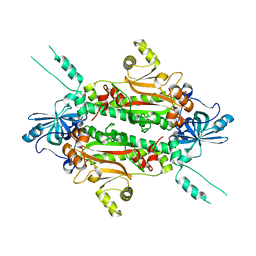 | |
6FC3
 
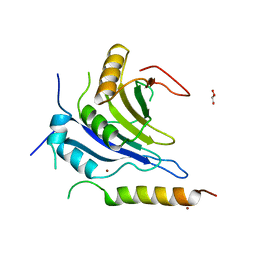 | |
7QPC
 
 | | Inward-facing NPA bound form of auxin transporter PIN8 | | Descriptor: | 1,2-DILINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 8 | | Authors: | Ung, K.L, Winkler, M.B.L, Dedic, E, Stokes, D.L, Pedersen, B.P. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-06 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structures and mechanism of the plant PIN-FORMED auxin transporter.
Nature, 609, 2022
|
|
8UX1
 
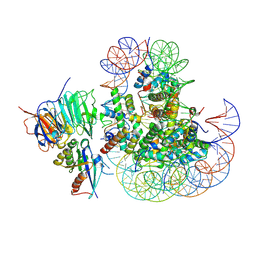 | |
6V5P
 
 | | EGFR(T790M/V948R) in complex with LN2725 | | Descriptor: | 4-[4-(4-fluorophenyl)-2-(3-methoxypropyl)-1H-imidazol-5-yl]-2-phenyl-3H-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2019-12-04 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for EGFR Mutant Inhibition by Trisubstituted Imidazole Inhibitors.
J.Med.Chem., 63, 2020
|
|
6YHT
 
 | | A lid blocking mechanism of a cone snail toxin revealed at the atomic level | | Descriptor: | CITRIC ACID, Conk-C1, SULFATE ION | | Authors: | Saikia, C, Altman-Gueta, H, Dym, O, Frolow, F, Gurevitz, M, Gordon, D, Reuveny, E, Karbat, I. | | Deposit date: | 2020-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Molecular Lid Mechanism of K + Channel Blocker Action Revealed by a Cone Peptide.
J.Mol.Biol., 433, 2021
|
|
6V6L
 
 | | Co-structure of human glycogen synthase kinase beta with 1-(6-((2-((6-amino-5-nitropyridin-2-yl)amino)ethyl)amino)-2-(2,4-dichlorophenyl)pyridin-3-yl)-4-methylpiperazin-2-one | | Descriptor: | 1-(6-((2-((6-amino-5-nitropyridin-2-yl)amino)ethyl)amino)-2-(2,4-dichlorophenyl)pyridin-3-yl)-4-methylpiperazin-2-one, Glycogen synthase kinase-3 beta, PHOSPHATE ION | | Authors: | Bussiere, D.E, Fang, E, Shu, W. | | Deposit date: | 2019-12-05 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery and optimization of novel pyridines as highly potent and selective glycogen synthase kinase 3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7ZYA
 
 | | Structure of Chit33 from Trichoderma harzianum. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Endochitinase 33, ... | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2022-05-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structure-Function Insights into the Fungal Endo -Chitinase Chit33 Depict its Mechanism on Chitinous Material.
Int J Mol Sci, 23, 2022
|
|
3IXY
 
 | | The pseudo-atomic structure of dengue immature virus in complex with Fab fragments of the anti-fusion loop antibody E53 | | Descriptor: | E53 Fab Fragment (chain H), E53 Fab Fragment (chain L), Envelope protein E, ... | | Authors: | Cherrier, M.V, Kaufmann, B, Nybakken, G.E, Lok, S.M, Warren, J.T, Nelson, C.A, Kostyuchenko, V.A, Holdaway, H.A, Chipman, P.R, Kuhn, R.J, Diamond, M.S, Rossmann, M.G, Fremont, D.H. | | Deposit date: | 2009-02-26 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody
Embo J., 28, 2009
|
|
6FFT
 
 | | Neutron structure of human transthyretin (TTR) S52P mutant in complex with tafamidis at room temperature to 2A resolution (quasi-Laue) | | Descriptor: | 2-(3,5-dichlorophenyl)-1,3-benzoxazole-6-carboxylic acid, Transthyretin | | Authors: | Yee, A.W, Moulin, M, Blakeley, M.P, Haertlein, M, Mitchell, E.P, Forsyth, V.T. | | Deposit date: | 2018-01-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | A molecular mechanism for transthyretin amyloidogenesis.
Nat Commun, 10, 2019
|
|
8A43
 
 | | Human RNA polymerase I | | Descriptor: | DNA-directed RNA polymerase I subunit RPA1, DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA2, ... | | Authors: | Daiss, J.L, Pilsl, M, Straub, K, Bleckmann, A, Hoecherl, M, Heiss, F.B, Abascal-Palacios, G, Ramsay, E, Tluckova, K, Mars, J.C, Fuertges, T, Bruckmann, A, Rudack, T, Bernecky, C, Lamour, V, Panov, K, Vannini, A, Moss, T, Engel, C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-31 | | Last modified: | 2022-11-02 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | The human RNA polymerase I structure reveals an HMG-like docking domain specific to metazoans.
Life Sci Alliance, 5, 2022
|
|
6YJ2
 
 | | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism | | Descriptor: | GLYCEROL, Probable transcriptional regulatory protein (Probably TetR-family) | | Authors: | Keep, N.H, Pritchard, J.E, Sula, A, Cole, A.R, Kendall, S.L. | | Deposit date: | 2020-04-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and DNA binding studies of the transcriptional repressor Rv2506 (BkaR) from Mycobacterium tuberculosis supports a role in L-Leucine catabolism
To be published
|
|
2J5D
 
 | | NMR structure of BNIP3 transmembrane domain in lipid bicelles | | Descriptor: | BCL2/ADENOVIRUS E1B 19 KDA PROTEIN-INTERACTING PROTEIN 3 | | Authors: | Bocharov, E.V, Pustovalova, Y.E, Volynsky, P.E, Maslennikov, I.V, Goncharuk, M.V, Ermolyuk, Y.S, Arseniev, A.S. | | Deposit date: | 2006-09-14 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Unique dimeric structure of BNip3 transmembrane domain suggests membrane permeabilization as a cell death trigger.
J. Biol. Chem., 282, 2007
|
|
1DTQ
 
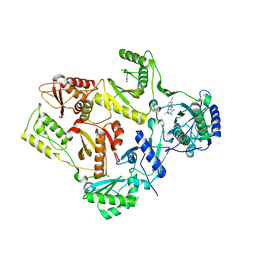 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH PETT-1 (PETT131A94) | | Descriptor: | HIV-1 RT A-CHAIN, HIV-1 RT B-CHAIN, N-[[3-FLUORO-4-ETHOXY-PYRID-2-YL]ETHYL]-N'-[5-NITRILOMETHYL-PYRIDYL]-THIOUREA | | Authors: | Ren, J, Diprose, J, Warren, J, Esnouf, R.M, Bird, L.E, Ikemizu, S, Slater, M, Milton, J, Balzarini, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2000-01-13 | | Release date: | 2000-03-20 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phenylethylthiazolylthiourea (PETT) non-nucleoside inhibitors of HIV-1 and HIV-2 reverse transcriptases. Structural and biochemical analyses.
J.Biol.Chem., 275, 2000
|
|
6BDX
 
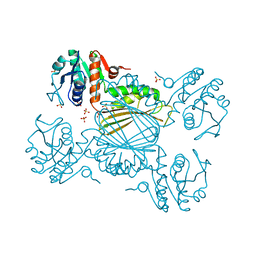 | | 4-hydroxy tetrahydrodipicolinate reductase from Neisseria gonorrhoeae | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, SULFATE ION | | Authors: | Pote, S.S, Pye, S.E, Sheahan, T.E, Chruszcz, M. | | Deposit date: | 2017-10-24 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 4-Hydroxy-tetrahydrodipicolinate reductase from Neisseria gonorrhoeae - structure and interactions with coenzymes and substrate analog.
Biochem. Biophys. Res. Commun., 503, 2018
|
|
6PLR
 
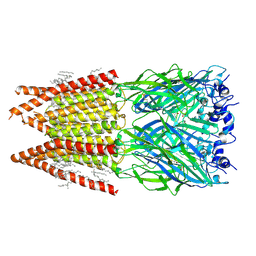 | | CryoEM structure of zebra fish alpha-1 glycine receptor bound with glycine in nanodisc, desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1, ... | | Authors: | Yu, J, Zhu, H, Gouaux, E. | | Deposit date: | 2019-07-01 | | Release date: | 2021-01-06 | | Last modified: | 2021-07-21 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of gating and partial agonist action in the glycine receptor.
Cell, 184, 2021
|
|
7QOA
 
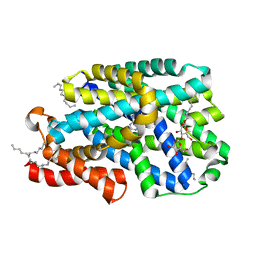 | | Structure of CodB, a cytosine transporter in an outward-facing conformation | | Descriptor: | 2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-AMINOPYRIMIDIN-2(1H)-ONE, Cytosine permease, ... | | Authors: | Hatton, C.E, Cameron, A.D. | | Deposit date: | 2021-12-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of cytosine transport protein CodB provides insight into nucleobase-cation symporter 1 mechanism.
Embo J., 41, 2022
|
|
1PU0
 
 | | Structure of Human Cu,Zn Superoxide Dismutase | | Descriptor: | COPPER (I) ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | DiDonato, M, Craig, L, Huff, M.E, Thayer, M.M, Cardoso, R.M.F, Kassmann, C.J, Lo, T.P, Bruns, C.K, Powers, E.T, Kelly, J.W, Getzoff, E.D, Tainer, J.A. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ALS Mutants of Human Superoxide Dismutase Form Fibrous Aggregates Via Framework Destabilization
J.Mol.Biol., 332, 2003
|
|
5AG2
 
 | | SOD-3 azide complex | | Descriptor: | ACETATE ION, AZIDE ION, MALONATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Bonetta, R, Stewart, E.E, Cabelli, D.E, Hunter, T. | | Deposit date: | 2015-01-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Structure of the Caenorhabditis Elegans Manganese Superoxide Dismutase Mnsod-3-Azide Complex.
Protein Sci., 24, 2015
|
|
8UQN
 
 | | PLCb3-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
