7ZLJ
 
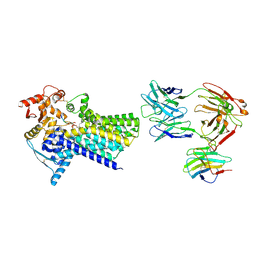 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
7ZLI
 
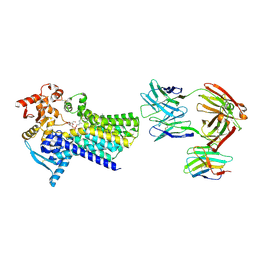 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in complex with Dol25-P-Man and bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
6VN7
 
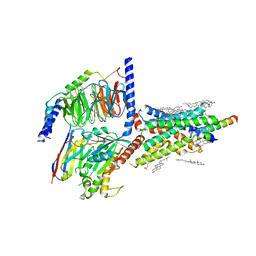 | | Cryo-EM structure of an activated VIP1 receptor-G protein complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Duan, J, Shen, D.-D, Zhou, X.E, Liu, Q.-F, Zhuang, Y.-W, Zhang, H.-B, Xu, P.-Y, Ma, S.-S, He, X.-H, Melcher, K, Zhang, Y, Xu, H.E, Yi, J. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of an activated VIP1 receptor-G protein complex revealed by a NanoBiT tethering strategy.
Nat Commun, 11, 2020
|
|
6PQB
 
 | |
5MHQ
 
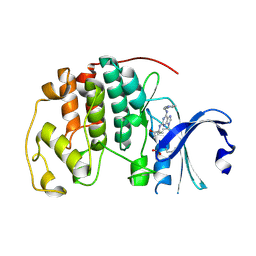 | | CCT068127 in complex with CDK2 | | Descriptor: | (2~{R},3~{S})-3-[[9-propan-2-yl-6-(pyridin-3-ylmethylamino)purin-2-yl]amino]pentan-2-ol, Cyclin-dependent kinase 2 | | Authors: | Whittaker, S.R, Barlow, C, Martin, M.P, Mancusi, C, Wagner, S, Barrie, E, te Poele, R, Sharp, S, Brown, N, Wilson, S, Clarke, P, Walton, M.I, MacDonald, E, Blagg, J, Noble, M.E.M, Garrett, M.D, Workman, P. | | Deposit date: | 2016-11-25 | | Release date: | 2017-12-20 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular profiling and combinatorial activity of CCT068127: a potent CDK2 and CDK9 inhibitor.
Mol Oncol, 12, 2018
|
|
4WZV
 
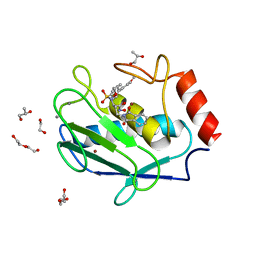 | | Crystal structure of a hydroxamate based inhibitor EN140 in complex with the MMP-9 catalytic domain | | Descriptor: | (2R)-4-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)-N-hydroxy-2-{[(4'-methoxybiphenyl-4-yl)sulfonyl](propan-2-yloxy)amino}butanamide, 1,2-ETHANEDIOL, AZIDE ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Dive, V, Rossello, A. | | Deposit date: | 2014-11-20 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | N-O-Isopropyl Sulfonamido-Based Hydroxamates as Matrix Metalloproteinase Inhibitors: Hit Selection and in Vivo Antiangiogenic Activity.
J.Med.Chem., 58, 2015
|
|
7BE7
 
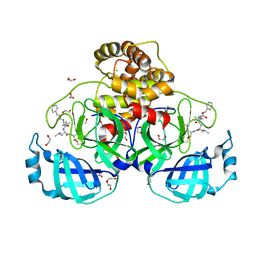 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7BGP
 
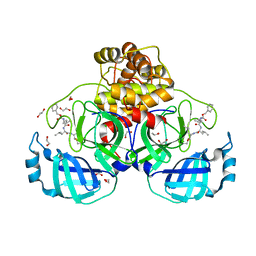 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in absence of DTT. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7UFX
 
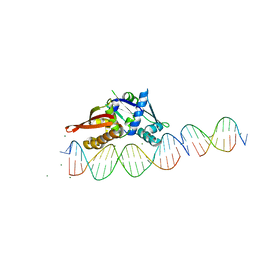 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and scaffold-insert duplexes (21mer and 10mer) containing the cognate REPE54 sequence and an additional G-C rich sequence, respectively. | | Descriptor: | DNA (5'-D(A*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*A)-3'), DNA (5'-D(A*GP*AP*CP*GP*GP*CP*CP*CP*GP*G)-3'), DNA (5'-D(A*GP*TP*CP*CP*GP*GP*GP*CP*CP*G)-3'), ... | | Authors: | Ward, A.R, Shields, E.T, Snow, C.D. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modular Protein-DNA Cocrystals as Precise, Programmable Assembly Scaffolds.
Acs Nano, 17, 2023
|
|
6NW0
 
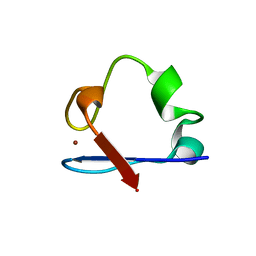 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
8U46
 
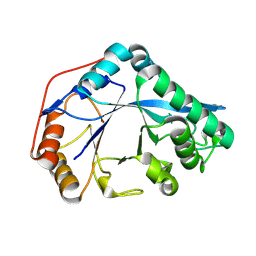 | |
6GFI
 
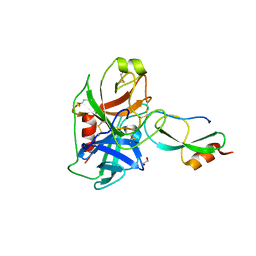 | | Structure of Human Mesotrypsin in complex with APPI variant T11V/M17R/I18F/F34V | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, PRSS3 protein | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N, Naftaly, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries.
Nat Commun, 9, 2018
|
|
6VZS
 
 | | Engineered TTLL6 mutant bound to gamma-elongation analog | | Descriptor: | (2~{S})-2-[[[(3~{R})-3-acetamido-4-oxidanyl-4-oxidanylidene-butyl]-phosphonooxy-phosphoryl]methyl]pentanedioic acid, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Mahalingan, K.K, Keenen, E.K, Strickland, M, Li, Y, Liu, Y, Ball, H.L, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for polyglutamate chain initiation and elongation by TTLL family enzymes.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Q3Z
 
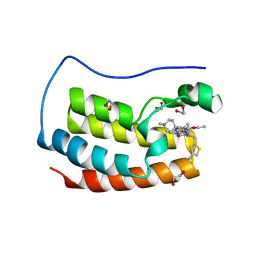 | | Crystal structure of the first bromodomain of human BRD4 in complex with the inhibitor 16k | | Descriptor: | (7~{R})-2-[[2-ethoxy-4-(1-methylpiperidin-4-yl)phenyl]amino]-7-ethyl-5-methyl-8-[(4-methylthiophen-2-yl)methyl]-7~{H}-pteridin-6-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Heidenreich, D, Watts, E, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Knapp, S, Hoelder, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing Dual Inhibitors of Anaplastic Lymphoma Kinase (ALK) and Bromodomain-4 (BRD4) by Tuning Kinase Selectivity.
J.Med.Chem., 62, 2019
|
|
6Q4J
 
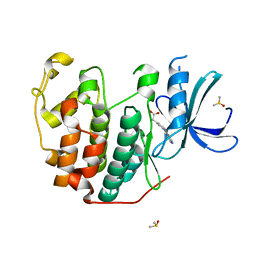 | | CDK2 in complex with FragLite34 | | Descriptor: | 2-[3-(pyrimidin-4-ylamino)phenyl]ethanoic acid, Cyclin-dependent kinase 2, DIMETHYL SULFOXIDE | | Authors: | Wood, D.J, Martin, M.P, Noble, M.E.M. | | Deposit date: | 2018-12-05 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | FragLites-Minimal, Halogenated Fragments Displaying Pharmacophore Doublets. An Efficient Approach to Druggability Assessment and Hit Generation.
J.Med.Chem., 62, 2019
|
|
6NK8
 
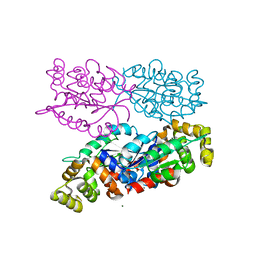 | | C-terminal region of the Burkholderia pseudomallei OLD protein | | Descriptor: | Class 2 OLD family nuclease, MAGNESIUM ION | | Authors: | Schiltz, C.J, Lee, A, Partlow, E.A, Hosford, C.J, Chappie, J.S. | | Deposit date: | 2019-01-05 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural characterization of Class 2 OLD family nucleases supports a two-metal catalysis mechanism for cleavage.
Nucleic Acids Res., 47, 2019
|
|
4Y8G
 
 | | Yeast 20S proteasome in complex with N3-APnLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6GXT
 
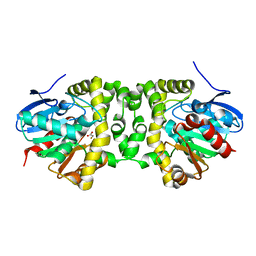 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography: FAcD2052MS after reaction initiation | | Descriptor: | Fluoroacetate dehalogenase, fluoroacetic acid | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-Werkmeister, H, Tellkamp, F, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E.F, Miller, R.J.D. | | Deposit date: | 2018-06-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
6CI9
 
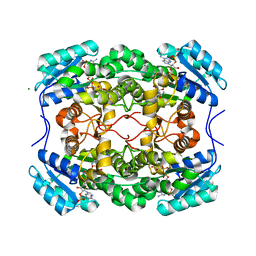 | |
8OKL
 
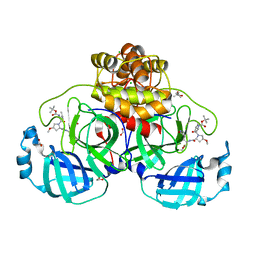 | | Crystal structure of F2F-2020185-01X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
8OKK
 
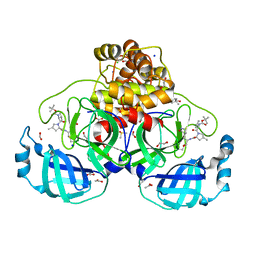 | | Crystal structure of F2F-2020184-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
6PWN
 
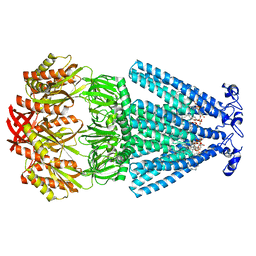 | | MscS Nanodisc with N-terminal His-Tag | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, HEXADECANE, Small-conductance mechanosensitive channel | | Authors: | Reddy, B.G, Perozo, E. | | Deposit date: | 2019-07-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of force-from-lipids gating in the mechanosensitive channel MscS.
Elife, 8, 2019
|
|
8OKM
 
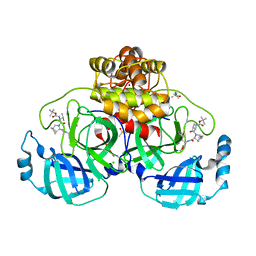 | | Crystal structure of F2F-2020197-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
6NPS
 
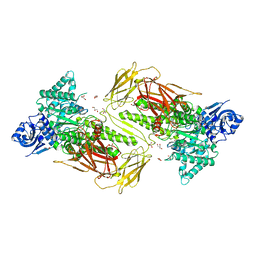 | | Crystal structure of GH115 enzyme AxyAgu115A from Amphibacillus xylanus | | Descriptor: | AxyAgu115A, CHLORIDE ION, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Yan, R, Master, E, Savchenko, A. | | Deposit date: | 2019-01-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural characterization of the family GH115 alpha-glucuronidase from Amphibacillus xylanus yields insight into its coordinated action with alpha-arabinofuranosidases.
N Biotechnol, 2021
|
|
8OKN
 
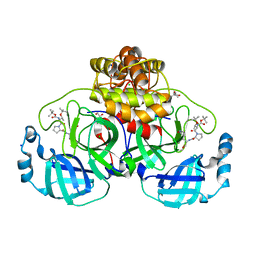 | | Crystal structure of F2F-2020198-00X bound to the main protease (3CLpro/Mpro) of SARS-CoV-2. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-03-28 | | Release date: | 2023-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination.
Eur.J.Med.Chem., 253, 2023
|
|
