2MFF
 
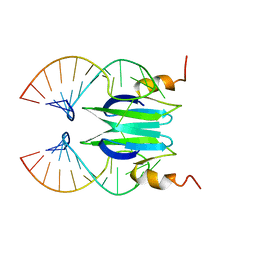 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL3)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL3(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFH
 
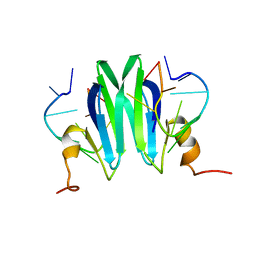 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(36-44)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, RsmZ(36-44) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFE
 
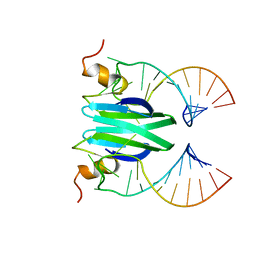 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL2)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL2(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFC
 
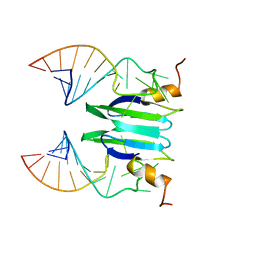 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL1)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL1(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-10 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MFG
 
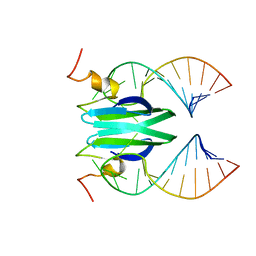 | | Csr/Rsm protein-RNA recognition - A molecular affinity ruler: RsmZ(SL4)/RsmE(dimer) 2:1 complex | | Descriptor: | Carbon storage regulator homolog, SL4(RsmZ) RNA | | Authors: | Duss, O, Diarra Dit Konte, N, Michel, E, Schubert, M, Allain, F.H.-T. | | Deposit date: | 2013-10-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for the wide range of affinity found in Csr/Rsm protein-RNA recognition.
Nucleic Acids Res., 42, 2014
|
|
2MF0
 
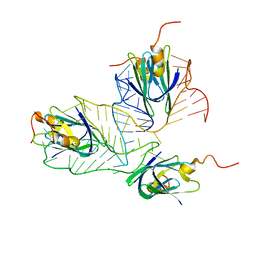 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer L of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2MF1
 
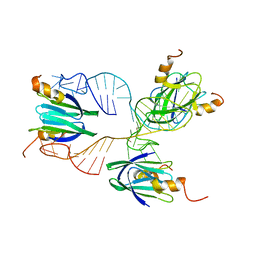 | | Structural basis of the non-coding RNA RsmZ acting as protein sponge: Conformer R of RsmZ(1-72)/RsmE(dimer) 1to3 complex | | Descriptor: | Carbon storage regulator homolog, RNA_(72-MER) | | Authors: | Duss, O, Michel, E, Yulikov, M, Schubert, M, Jeschke, G, Allain, F.H.-T. | | Deposit date: | 2013-10-02 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of the non-coding RNA RsmZ acting as a protein sponge.
Nature, 509, 2014
|
|
2JPP
 
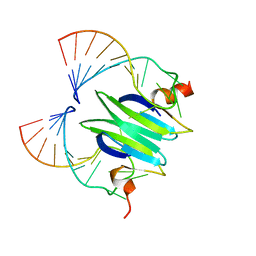 | | Structural basis of RsmA/CsrA RNA recognition: Structure of RsmE bound to the Shine-Dalgarno sequence of hcnA mRNA | | Descriptor: | RNA (5'-R(*GP*GP*GP*CP*UP*UP*CP*AP*CP*GP*GP*AP*UP*GP*AP*AP*GP*CP*CP*C)-3'), Translational repressor | | Authors: | Schubert, M, Lapouge, K, Duss, O, Oberstrass, F.C, Jelesarov, I, Haas, D, Allain, F.H.-T. | | Deposit date: | 2007-05-21 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of messenger RNA recognition by the specific bacterial repressing clamp RsmA/CsrA
Nat.Struct.Mol.Biol., 14, 2007
|
|
3V80
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-O-Propargylamino-5'-deoxyadenosine | | Descriptor: | 5'-O-prop-2-yn-1-yladenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.0301 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V7U
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with MTA | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8N
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 8-bromo-5'-amino-5'-deoxyadenosine, reacted with a citrate molecule in N site | | Descriptor: | 5'-amino-8-bromo-5'-deoxyadenosine, 8-bromo-5'-{[3-carboxy-2-(carboxymethyl)-2-hydroxypropanoyl]amino}-5'-deoxyadenosine, CITRIC ACID, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3801 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8R
 
 | | Crystal structure of NAD kinase 1 H223E mutant from Listeria monocytogenes in complex with 5'-amino-8-bromo-5'-deoxyadenosine | | Descriptor: | 5'-amino-8-bromo-5'-deoxyadenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8P
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a new di-adenosine inhibitor formed in situ | | Descriptor: | 2-[6-azanyl-9-[(2R,3R,4S,5R)-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-3,4-bis(oxidanyl)oxolan-2-yl]purin-8-yl]sulfanyl-N-[[(2R,3S,4R,5R)-5-(6-azanyl-8-bromanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]ethanamide, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2901 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V7Y
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-N-Propargylamino-5'-deoxyadenosine | | Descriptor: | 5'-deoxy-5'-(prop-2-yn-1-ylamino)adenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V7W
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with 5'-azido-5'-deoxyadenosine | | Descriptor: | 5'-azido-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-22 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.0102 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8M
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complexe with 5'-azido-8-bromo-5'-deoxyadenosine | | Descriptor: | 5'-azido-8-bromo-5'-deoxyadenosine, CITRIC ACID, Probable inorganic polyphosphate/ATP-NAD kinase 1 | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
3V8Q
 
 | | Crystal structure of NAD kinase 1 H223E mutant from Listeria monocytogenes in complex with 5'-amino-5'-deoxyadenosine | | Descriptor: | 5'-amino-5'-deoxyadenosine, CITRIC ACID, GLYCEROL, ... | | Authors: | Gelin, M, Poncet-Montange, G, Assairi, L, Morellato, L, Huteau, V, Dugu, L, Dussurget, O, Pochet, S, Labesse, G. | | Deposit date: | 2011-12-23 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Screening and In Situ Synthesis Using Crystals of a NAD Kinase Lead to a Potent Antistaphylococcal Compound.
Structure, 20, 2012
|
|
