3LDV
 
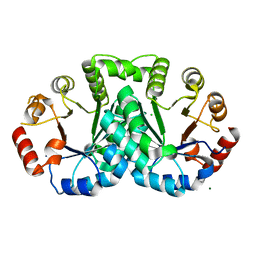 | | 1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Dubrovska, I, Winsor, J, Glass, E.M, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-13 | | Release date: | 2010-01-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | 1.77 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
5V36
 
 | | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-06 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD.
To Be Published
|
|
3LV8
 
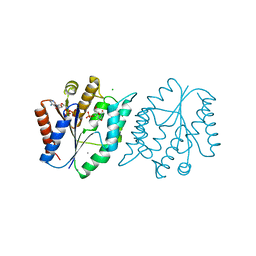 | | 1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Halavaty, A.S, Minasov, G, Dubrovska, I, Winsor, J, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-19 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of a thymidylate kinase (tmk) from Vibrio cholerae O1 biovar eltor str. N16961 in complex with TMP, thymidine-5'-diphosphate and ADP
To be Published
|
|
3LWZ
 
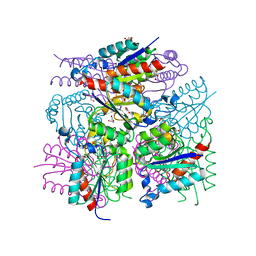 | | 1.65 Angstrom Resolution Crystal Structure of Type II 3-Dehydroquinate Dehydratase (aroQ) from Yersinia pestis | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate dehydratase, BETA-MERCAPTOETHANOL, ... | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1.65 Angstrom Resolution Crystal Structure of Type II 3-Dehydroquinate Dehydratase (aroQ) from Yersinia pestis.
TO BE PUBLISHED
|
|
3M07
 
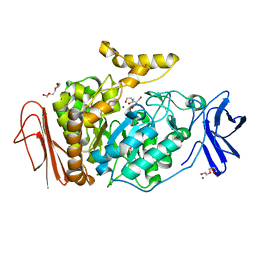 | | 1.4 Angstrom Resolution Crystal Structure of Putative alpha Amylase from Salmonella typhimurium. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 Angstrom Resolution Crystal Structure of Putative alpha Amylase from Salmonella typhimurium.
TO BE PUBLISHED
|
|
3M3H
 
 | | 1.75 Angstrom resolution crystal structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | CHLORIDE ION, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Shuvalova, L, Minasov, G, Winsor, J, Dubrovska, I, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-09 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 1.75 Angstrom resolution crystal structure of an orotate phosphoribosyltransferase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3MJD
 
 | | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Orotate phosphoribosyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Crystal Structure of Orotate Phosphoribosyltransferase (pyrE) Francisella tularensis.
TO BE PUBLISHED
|
|
3N2B
 
 | | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae. | | Descriptor: | CHLORIDE ION, Diaminopimelate decarboxylase | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Diaminopimelate Decarboxylase (lysA) from Vibrio cholerae.
To be Published
|
|
4LES
 
 | | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, ETHANOL, PHOSPHATE ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Filippova, E, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Conserved Hypothetical Protein from Bacillus anthracis.
TO BE PUBLISHED
|
|
3N2L
 
 | | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase (pyrE) from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | CHLORIDE ION, Orotate phosphoribosyltransferase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom resolution crystal structure of an Orotate Phosphoribosyltransferase
(pyrE) from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
5W6L
 
 | | Crystal Structure of RRSP, a MARTX Toxin Effector Domain from Vibrio vulnificus CMCP6 | | Descriptor: | CHLORIDE ION, GLYCEROL, RTX repeat-containing cytotoxin, ... | | Authors: | Minasov, G, Wawrzak, Z, Biancucci, M, Shuvalova, L, Dubrovska, I, Satchell, K.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-06-16 | | Release date: | 2018-06-27 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | The bacterial Ras/Rap1 site-specific endopeptidase RRSP cleaves Ras through an atypical mechanism to disrupt Ras-ERK signaling.
Sci Signal, 11, 2018
|
|
4NV4
 
 | | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Signal peptidase I, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-04 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Crystal Structure of Signal Peptidase I from Bacillus anthracis.
TO BE PUBLISHED
|
|
3RU6
 
 | | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168 | | Descriptor: | CHLORIDE ION, IODIDE ION, Orotidine 5'-phosphate decarboxylase | | Authors: | Halavaty, A.S, Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-04 | | Release date: | 2011-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom resolution crystal structure of orotidine 5'-phosphate decarboxylase (pyrF) from Campylobacter jejuni subsp. jejuni NCTC 11168
To be Published
|
|
3SG1
 
 | | 2.6 Angstrom Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurA1) from Bacillus anthracis | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 | | Authors: | Minasov, G, Halavaty, A, Filippova, E.V, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase 1 (MurA1) from Bacillus anthracis.
TO BE PUBLISHED
|
|
5DZS
 
 | | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile. | | Descriptor: | SULFATE ION, Shikimate dehydrogenase (NADP(+)) | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Shikimate Dehydrogenase 1 from Peptoclostridium difficile.
To Be Published
|
|
5E31
 
 | | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium. | | Descriptor: | Penicillin binding protein 2 prime | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Flores, K, Filippova, E, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-10-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 2.3 Angstrom Crystal Structure of the Monomeric Form of Penicillin Binding Protein 2 Prime from Enterococcus faecium.
To Be Published
|
|
4MLG
 
 | | Structure of RS223-Beta-xylosidase | | Descriptor: | Beta-xylosidase, CALCIUM ION, SULFATE ION | | Authors: | Jordan, D, Braker, J, Wagschal, K, Lee, C, Dubrovska, I, Anderson, S, Wawrzak, Z. | | Deposit date: | 2013-09-06 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of RS223-Beta-xylosidase
To be Published
|
|
5T1Q
 
 | | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus. | | Descriptor: | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979, SODIUM ION, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bagnoli, F, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 2.15 Angstrom Crystal Structure of N-acetylmuramoyl-L-alanine Amidase from Staphylococcus aureus.
To Be Published
|
|
3K96
 
 | | 2.1 Angstrom resolution crystal structure of glycerol-3-phosphate dehydrogenase (gpsA) from Coxiella burnetii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, Glycerol-3-phosphate dehydrogenase [NAD(P)+] | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-10-15 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2.1 Angstrom Resolution Crystal Structure of Glycerol-3-phosphate Dehydrogenase (gpsA) from Coxiella burnetii.
TO BE PUBLISHED
|
|
5T8K
 
 | | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-07 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Adenine and NAD.
To Be Published
|
|
5TJ9
 
 | | 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD | | Descriptor: | (1R,2S,3R,5R)-3-(6-amino-9H-purin-9-yl)-5-(hydroxymethyl)cyclopentane-1,2-diol, Adenosylhomocysteinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-04 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2.6 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with Aristeromycin and NAD
To Be Published
|
|
5TLS
 
 | | 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD | | Descriptor: | (2S)-4-(6-amino-9H-purin-9-yl)-2-hydroxybutanoic acid, Adenosylhomocysteinase, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Bishop, B, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-11 | | Release date: | 2016-10-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 Angstrom Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with DZ2002 and NAD
To Be Published
|
|
5HJ1
 
 | | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid | | Descriptor: | Pullulanase C protein, VACCENIC ACID | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-12 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of PDZ domain of pullulanase C protein of type II secretion system from Klebsiella pneumoniae in complex with fatty acid
To Be Published
|
|
5HQH
 
 | | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes. | | Descriptor: | CHLORIDE ION, Lmo2119 protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | 1.32 Angstrom Crystal Structure of Ybbr like Domain of lmo2119 Protein from Listeria monocytogenes.
To Be Published
|
|
5U2G
 
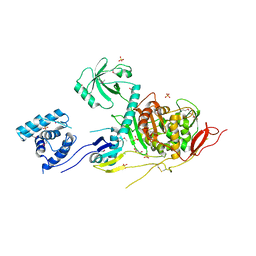 | | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | 2.6 Angstrom Resolution Crystal Structure of Penicillin-Binding Protein 1A from Haemophilus influenzae.
To Be Published
|
|
