9ISZ
 
 | |
9IT4
 
 | | Structure of Clr4 catalyzing histone H3 K9 methylation | | Descriptor: | Histone H3.1/H3.2, Histone-lysine N-methyltransferase, H3 lysine-9 specific, ... | | Authors: | Du, Y.X, Liu, L. | | Deposit date: | 2024-07-19 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Mechanism of Histone H3K9 Methyltransferase Clr4 Regulation by Proximal H3K14 Ubiquitination in-cis
To Be Published
|
|
6P29
 
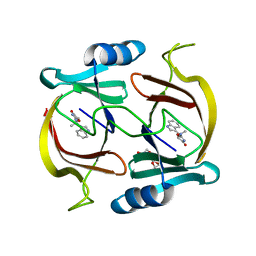 | | N-demethylindolmycin synthase (PluN2) in complex with N-demethylindolmycin | | Descriptor: | (5S)-2-amino-5-[(1R)-1-(1H-indol-3-yl)ethyl]-1,3-oxazol-4(5H)-one, N-demethylindolmycin synthase (PluN2), TRIETHYLENE GLYCOL | | Authors: | Du, Y.L, Higgins, M.A, Zhao, G, Ryan, K.S. | | Deposit date: | 2019-05-21 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Convergent biosynthetic transformations to a bacterial specialized metabolite.
Nat.Chem.Biol., 15, 2019
|
|
6LQU
 
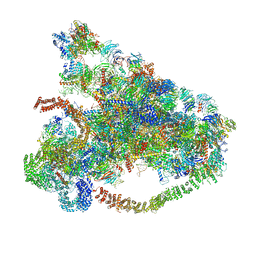 | |
8IYA
 
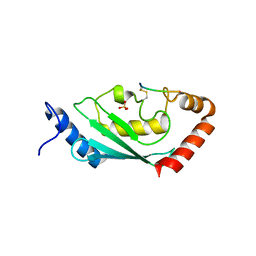 | | Complex of SETDB1-derived peptide bound to UBE2E1 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, SULFATE ION, Ubiquitin-conjugating enzyme E2 E1 | | Authors: | Du, Y.X, Liu, L. | | Deposit date: | 2023-04-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure-guided engineering enables E3 ligase-free and versatile protein ubiquitination via UBE2E1.
Nat Commun, 15, 2024
|
|
5YZ4
 
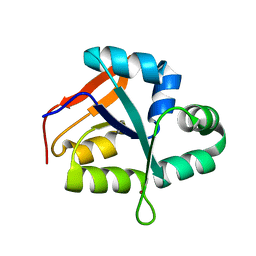 | | Structure of the PIN domain endonuclease Utp24 | | Descriptor: | CALCIUM ION, ZINC ION, rRNA-processing protein fcf1 | | Authors: | Du, Y, An, W, Ye, K. | | Deposit date: | 2017-12-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.135 Å) | | Cite: | Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA.
Plos One, 13, 2018
|
|
8JTL
 
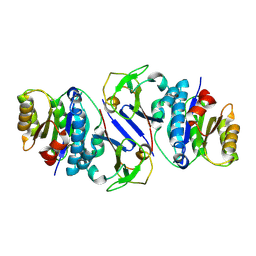 | | Structure of OY phytoplasma SAP05 binding with AtRpn10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
3LZZ
 
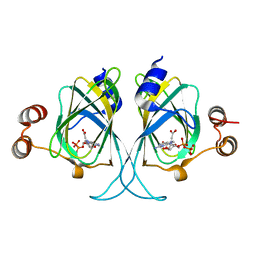 | | Crystal structures of Cupin superfamily BbDUF985 from Branchiostoma belcheri tsingtauense in apo and GDP-bound forms | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | Authors: | Du, Y, He, Y.-X, Saren, G, Zhang, X, Zhang, S.-C, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-03-02 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the apo and GDP-bound forms of a cupin-like protein BbDUF985 from Branchiostoma belcheri tsingtauense
Proteins, 2010
|
|
8JTK
 
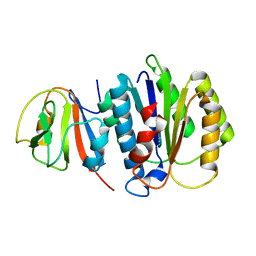 | | Structure of AYWB phytoplasma SAP05 recognizing AtRpn10 | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 4 homolog, Sequence-variable mosaic (SVM) signal sequence domain-containing protein | | Authors: | Du, Y.X, Zhang, L.Y, Zheng, Q.Y. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure basis for recognition of plant Rpn10 by phytoplasma SAP05 in ubiquitin-independent protein degradation.
Iscience, 27, 2024
|
|
3NZ3
 
 | | Crystal structure of the mucin-binding domain of Spr1345 from Streptococcus pneumoniae | | Descriptor: | Putative uncharacterized protein, SULFATE ION, TRIETHYLENE GLYCOL | | Authors: | Du, Y, He, Y.-X, Zhang, Z.-Y, Yang, Y.-H, Shi, W.-W, Frolet, C, Guilmi, A.M, Vernet, T, Zhou, C.-Z, Chen, Y. | | Deposit date: | 2010-07-15 | | Release date: | 2011-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the mucin-binding domain of Spr1345 from Streptococcus pneumoniae
J.Struct.Biol., 174, 2011
|
|
3PPP
 
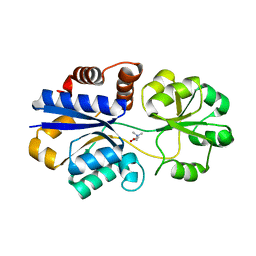 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein, TRIMETHYL GLYCINE | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPO
 
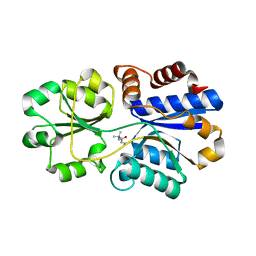 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (2S)-3-carboxy-2-hydroxy-N,N,N-trimethylpropan-1-aminium, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPN
 
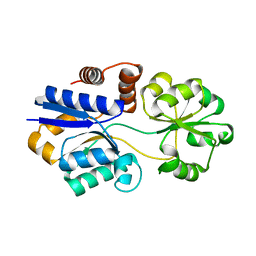 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPQ
 
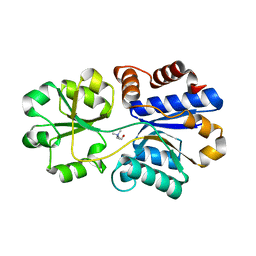 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | CHOLINE ION, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3PPR
 
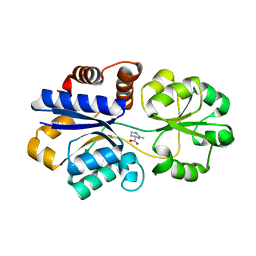 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
6KE6
 
 | |
6LQS
 
 | |
6LQR
 
 | |
6LQV
 
 | |
6LQT
 
 | |
6LQQ
 
 | |
6LQP
 
 | |
8WKC
 
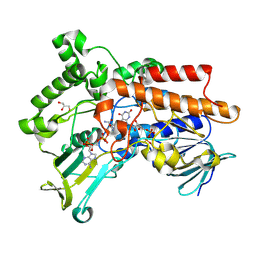 | | Crystal structure of OgBVMO(Oceanicola granulosus) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Du, Y, Wang, Y.H. | | Deposit date: | 2023-09-27 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of OgBVMO(Oceanicola granulosus)
To be published
|
|
7D63
 
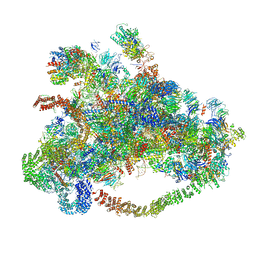 | | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-29 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | Cryo-EM structure of 90S preribosome with inactive Utp24 (state C)
To Be Published
|
|
7D4I
 
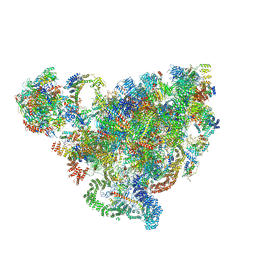 | | Cryo-EM structure of 90S small ribosomal precursors complex with the DEAH-box RNA helicase Dhr1 (State F) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Du, Y, Zhang, J, An, W, Ye, K. | | Deposit date: | 2020-09-24 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of 90S small ribosomal precursors complex with Dhr1
To Be Published
|
|
