1G87
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDOCELLULASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-16 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1GZ1
 
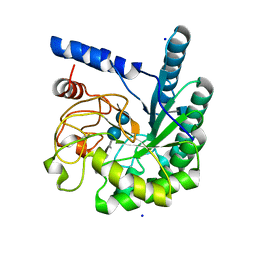 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with methyl-cellobiosyl-4-deoxy-4-thio-beta-D-cellobioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, SODIUM ION, ... | | Authors: | Varrot, A, Frandsen, T, Driguez, H, Davies, G.J. | | Deposit date: | 2002-05-13 | | Release date: | 2003-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Humicola Insolens Cellobiohydrolase Cel6A D416A Mutant in Complex with a Non-Hydrolysable Substrate Analogue, Methyl Cellobiosyl-4-Thio-Beta-Cellobioside, at 1.9 A
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H5V
 
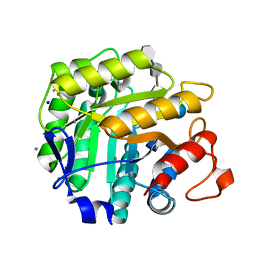 | | Thiopentasaccharide complex of the endoglucanase Cel5A from Bacillus agaradharens at 1.1 A resolution in the tetragonal crystal form | | Descriptor: | CALCIUM ION, ENDOGLUCANASE 5A, SODIUM ION, ... | | Authors: | Varrot, A, Sulzenbacher, G, Schulein, M, Driguez, H, Davies, G.J. | | Deposit date: | 2001-05-28 | | Release date: | 2002-05-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Structure of Endoglucanase Cel5A in Complex with Methyl 4,4II,4III,4Iv-Tetrathio-Alpha-Cellopentoside Highlights the Alternative Binding Modes Targeted by Substrate Mimics
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1GA2
 
 | | THE CRYSTAL STRUCTURE OF ENDOGLUCANASE 9G FROM CLOSTRIDIUM CELLULOLYTICUM COMPLEXED WITH CELLOBIOSE | | Descriptor: | ACETIC ACID, CALCIUM ION, ENDOGLUCANASE 9G, ... | | Authors: | Mandelman, D, Belaich, A, Belaich, J.P, Aghajari, N, Driguez, H, Haser, R. | | Deposit date: | 2000-11-29 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray Crystal Structure of the Multidomain Endoglucanase Cel9G from Clostridium
cellulolyticum Complexed with Natural and Synthetic Cello-Oligosaccharides
J.BACTERIOL., 185, 2003
|
|
1J8V
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4'-nitrophenyl 3I-thiolaminaritrioside | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4'-NITROPHENYL-S-(BETA-D-GLUCOPYRANOSYL)-(1-3)-(3-THIO-BETA-D-GLUCOPYRANOSYL)-(1-3)-BETA-D-GLUCOPYRANOSIDE, ... | | Authors: | Hrmova, M, De Gori, R, Smith, B.J, Fairweather, J.K, Driguez, H, Varghese, J.N, Fincher, G.B. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for broad substrate specificity in higher plant beta-D-glucan glucohydrolases.
Plant Cell, 14, 2002
|
|
1IEX
 
 | | Crystal structure of barley beta-D-glucan glucohydrolase isoenzyme Exo1 in complex with 4I,4III,4V-S-trithiocellohexaose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Hrmova, M, DeGori, R, Fincher, G.B, Smith, B.J, Driguez, H, Varghese, J.N. | | Deposit date: | 2001-04-11 | | Release date: | 2001-11-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanisms and reaction intermediates along the hydrolytic pathway of a plant beta-D-glucan glucohydrolase.
Structure, 9, 2001
|
|
1DYM
 
 | | Humicola insolens Endocellulase Cel7B (EG 1) E197A Mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOGLUCANASE I | | Authors: | Davies, G.J, Moraz, O, Driguez, H, Schulein, M. | | Deposit date: | 2000-02-03 | | Release date: | 2000-02-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate.
Biochem.J., 335 ( Pt 2), 1998
|
|
6YQ4
 
 | | Crystal structure of Fusobacterium nucleatum tannase | | Descriptor: | GLYCEROL, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Mancheno, J.M, Anguita, J, Rodriguez, H. | | Deposit date: | 2020-04-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | A structurally unique Fusobacterium nucleatum tannase provides detoxicant activity against gallotannins and pathogen resistance.
Microb Biotechnol, 15, 2022
|
|
2W37
 
 | | CRYSTAL STRUCTURE OF THE HEXAMERIC CATABOLIC ORNITHINE TRANSCARBAMYLASE FROM Lactobacillus hilgardii | | Descriptor: | NICKEL (II) ION, ORNITHINE CARBAMOYLTRANSFERASE, CATABOLIC | | Authors: | de las Rivas, B, Fox, G.C, Angulo, I, Rodriguez, H, Munoz, R, Mancheno, J.M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Hexameric Catabolic Ornithine Transcarbamylase from Lactobacillus Hilgardii: Structural Insights Into the Oligomeric Assembly and Metal Binding.
J.Mol.Biol., 393, 2009
|
|
