7WDE
 
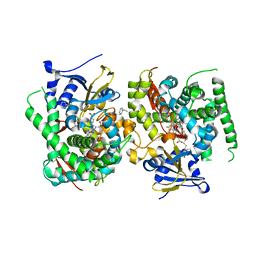 | | Crystal structure of the P450 BM3 heme domain mutant F87L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, GLYCEROL, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDI
 
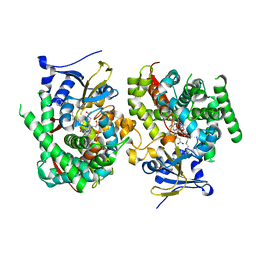 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
7WDD
 
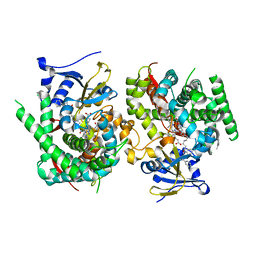 | | Crystal structure of the P450 BM3 heme domain mutant F87K in complex with N-imidazolyl-hexanoyl-L-phenylalanine, styrene and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanine and hydroxylamine
To Be Published
|
|
6L2S
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_D83G | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2R
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_E195S | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)) | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-06 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
1KHY
 
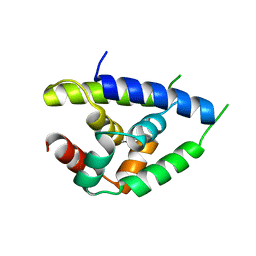 | |
1JBK
 
 | |
3QD2
 
 | | Crystal structure of mouse PERK kinase domain | | Descriptor: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Wenjun, C, Jingzhi, L, David, R, Bingdong, S. | | Deposit date: | 2011-01-17 | | Release date: | 2011-04-27 | | Last modified: | 2019-01-16 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of the PERK kinase domain suggests the mechanism for its activation.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6L2K
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_R49E | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), SULFATE ION | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2Z
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pnuemoniae_D191G | | Descriptor: | AMMONIUM ION, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-07 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
6L2I
 
 | | IlvC, a ketol-acid reductoisomerase, from Streptococcus pneumoniae_WT | | Descriptor: | GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, ... | | Authors: | Gyuhee, K, Donghyuk, S, Sumin, L, Jaesook, Y, Sangho, L. | | Deposit date: | 2019-10-04 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystal Structure of IlvC, a Ketol-Acid Reductoisomerase, from Streptococcus Pneumoniae.
Crystals, 9, 2019
|
|
