7ADQ
 
 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
2NDO
 
 | | Structure of EcDsbA-sulfonamide1 complex | | Descriptor: | 2-{[(4-iodophenyl)sulfonyl]amino}benzoic acid, Thiol:disulfide interchange protein DsbA | | Authors: | Williams, M.L, Doak, B.C, Vazirani, M, Ilyichova, O, Wang, G, Bermel, W, Simpson, J.S, Chalmers, D.K, King, G.F, Mobli, M, Scanlon, M.J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Determination of ligand binding modes in weak protein-ligand complexes using sparse NMR data.
J.Biomol.Nmr, 66, 2016
|
|
6DO6
 
 | | NMR solution structure of wild type apo hFABP1 at 308 K | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6DRG
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
6DO7
 
 | | NMR solution structure of wild type hFABP1 with GW7647 | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Scanlon, M.J, Mohanty, B, Doak, B.C, Patil, R. | | Deposit date: | 2018-06-09 | | Release date: | 2019-01-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A ligand-induced structural change in fatty acid-binding protein 1 is associated with potentiation of peroxisome proliferator-activated receptor alpha agonists.
J. Biol. Chem., 294, 2019
|
|
4WF5
 
 | | Crystal structure of E.Coli DsbA soaked with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, COPPER (II) ION, ... | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WEY
 
 | | Crystal structure of E.Coli DsbA in complex with compound 17 | | Descriptor: | 1,2-ETHANEDIOL, N-({4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazol-5-yl}carbonyl)-L-serine, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4WF4
 
 | | Crystal structure of E.Coli DsbA co-crystallised in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-2-[4-(trifluoromethyl)phenyl]-1,3-thiazole-5-carboxylic acid, Thiol:disulfide interchange protein | | Authors: | Adams, L.A, Sharma, P, Mohanty, B, Ilyichova, O.V, Mulcair, M.D, Williams, M.L, Gleeson, E.C, Totsika, M, Doak, B.C, Caria, S, Rimmer, K, Shouldice, S.R, Vazirani, M, Headey, S.J, Plumb, B.R, Martin, J.L, Heras, B, Simpson, J.S, Scanlon, M.J. | | Deposit date: | 2014-09-12 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Application of Fragment-Based Screening to the Design of Inhibitors of Escherichia coli DsbA.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5QKC
 
 | |
5QKS
 
 | |
5QL9
 
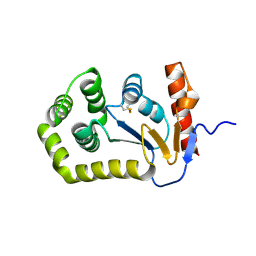 | |
5QLS
 
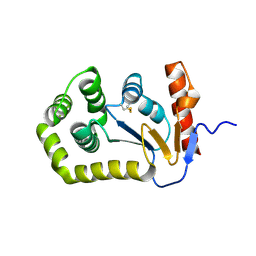 | |
5QM7
 
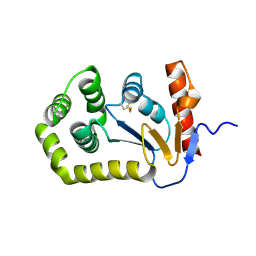 | |
5QKI
 
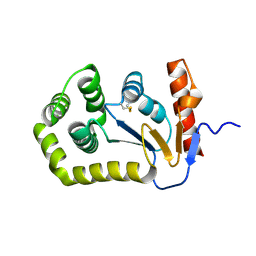 | |
5QKT
 
 | |
5QLE
 
 | |
5QLR
 
 | |
5QM3
 
 | |
5QMH
 
 | |
5QMY
 
 | |
5QKM
 
 | |
5QL1
 
 | |
5QLK
 
 | |
5QLZ
 
 | |
5QKK
 
 | |
