1AQ6
 
 | | STRUCTURE OF L-2-HALOACID DEHALOGENASE FROM XANTHOBACTER AUTOTROPHICUS | | Descriptor: | FORMIC ACID, L-2-HALOACID DEHALOGENASE | | Authors: | Ridder, I.S, Rozeboom, H.J, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 1997-08-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate.
J.Biol.Chem., 272, 1997
|
|
1EDD
 
 | |
1EDE
 
 | |
1ILZ
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 6.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1S0Y
 
 | | The structure of trans-3-chloroacrylic acid dehalogenase, covalently inactivated by the mechanism-based inhibitor 3-bromopropiolate at 2.3 Angstrom resolution | | Descriptor: | MALONIC ACID, alpha-subunit of trans-3-chloroacrylic acid dehalogenase, beta-subunit of trans-3-chloroacrylic acid dehalogenase | | Authors: | de Jong, R.M, Brugman, W, Poelarends, G.J, Whitman, C.P, Dijkstra, B.W. | | Deposit date: | 2004-01-05 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The X-ray structure of trans-3-chloroacrylic acid dehalogenase reveals a novel hydration mechanism in the tautomerase superfamily
J.Biol.Chem., 279, 2004
|
|
1RY9
 
 | | Spa15, a Type III Secretion Chaperone from Shigella flexneri | | Descriptor: | CHLORIDE ION, Surface presentation of antigens protein spaK | | Authors: | van Eerde, A, Hamiaux, C, Perez, J, Parsot, C, Dijkstra, B.W. | | Deposit date: | 2003-12-20 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure of Spa15, a type III secretion chaperone from Shigella flexneri with broad specificity.
Embo Rep., 5, 2004
|
|
1IIB
 
 | | CRYSTAL STRUCTURE OF IIBCELLOBIOSE FROM ESCHERICHIA COLI | | Descriptor: | ENZYME IIB OF THE CELLOBIOSE-SPECIFIC PHOSPHOTRANSFERASE SYSTEM | | Authors: | Van Montfort, R.L.M, Pijning, T, Kalk, K.H, Reizer, J, Saier, M.H, Thunnissen, M.M.G.M, Robillard, G.T, Dijkstra, B.W. | | Deposit date: | 1996-12-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of an energy-coupling protein from bacteria, IIBcellobiose, reveals similarity to eukaryotic protein tyrosine phosphatases.
Structure, 5, 1997
|
|
1CRU
 
 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS IN COMPLEX WITH PQQ AND METHYLHYDRAZINE | | Descriptor: | CALCIUM ION, GLYCEROL, METHYLHYDRAZINE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-16 | | Release date: | 2000-03-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4R9L
 
 | | Structure of a thermostable elevenfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis, containing two stabilizing disulfide bonds | | Descriptor: | (2R)-2-hydroxyhexanamide, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
4R9K
 
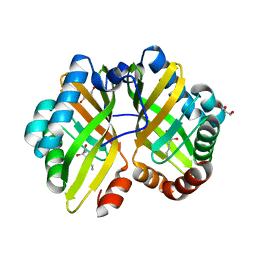 | | Structure of thermostable eightfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis | | Descriptor: | (2R)-2-hydroxyhexanamide, GLYCEROL, Limonene-1,2-epoxide hydrolase | | Authors: | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2014-09-05 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
5Z5H
 
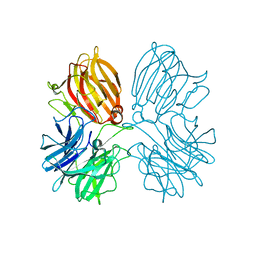 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5Z5F
 
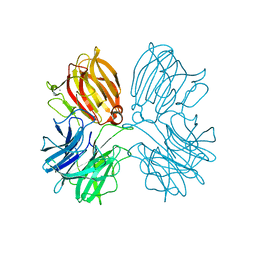 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose | | Descriptor: | Beta-xylosidase, CALCIUM ION, beta-L-arabinofuranose | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5Z5D
 
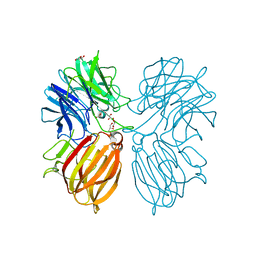 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 | | Descriptor: | Beta-xylosidase, CALCIUM ION, GLYCEROL | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-17 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
5Z5I
 
 | | Crystal structure of a thermostable glycoside hydrolase family 43 {beta}-1,4-xylosidase from Geobacillus thermoleovorans IT-08 in complex with L-arabinose and D-xylose | | Descriptor: | Beta-xylosidase, CALCIUM ION, alpha-D-xylopyranose, ... | | Authors: | Rohman, A, van Oosterwijk, N, Puspaningsih, N.N.T, Dijkstra, B.W. | | Deposit date: | 2018-01-18 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 beta-1,4-xylosidase from Geobacillus thermoleovorans IT-08.
PLoS ONE, 13, 2018
|
|
1CQ1
 
 | | Soluble Quinoprotein Glucose Dehydrogenase from Acinetobacter Calcoaceticus in Complex with PQQH2 and Glucose | | Descriptor: | CALCIUM ION, PYRROLOQUINOLINE QUINONE, SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-04 | | Release date: | 2000-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of soluble quinoprotein glucose dehydrogenase.
EMBO J., 18, 1999
|
|
3KLK
 
 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180 in triclinic apo- form | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5JBD
 
 | | 4,6-alpha-glucanotransferase GTFB from Lactobacillus reuteri 121 | | Descriptor: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
5JBF
 
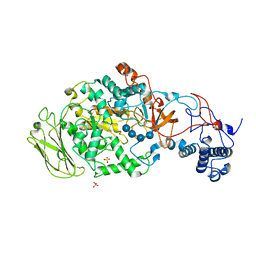 | | 4,6-alpha-glucanotransferase GTFB (D1015N mutant) from Lactobacillus reuteri 121 complexed with maltopentaose | | Descriptor: | CALCIUM ION, Inactive glucansucrase, SULFATE ION, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
5JBE
 
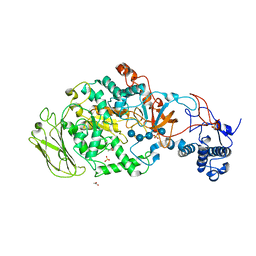 | | 4,6-alpha-glucanotransferase GTFB from Lactobacillus reuteri 121 complexed with an isomalto-maltopentasaccharide | | Descriptor: | ACETATE ION, CALCIUM ION, Inactive glucansucrase, ... | | Authors: | Pijning, T, Dijkstra, B.W, Bai, Y, Gangoiti-Munecas, J, Dijkhuizen, L. | | Deposit date: | 2016-04-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of 4,6-alpha-Glucanotransferase Supports Diet-Driven Evolution of GH70 Enzymes from alpha-Amylases in Oral Bacteria.
Structure, 25, 2017
|
|
3JUR
 
 | | The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima | | Descriptor: | Exo-poly-alpha-D-galacturonosidase | | Authors: | Pijning, T, van Pouderoyen, G, Kluskens, L.D, van der Oost, J, Dijkstra, B.W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of a hyperthermoactive exopolygalacturonase from Thermotoga maritima reveals a unique tetramer
Febs Lett., 2009
|
|
3KLL
 
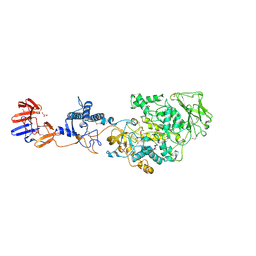 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180-maltose complex | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase, ... | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4AT2
 
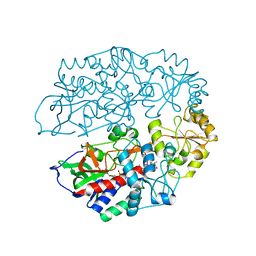 | | The crystal structure of 3-ketosteroid-delta4-(5alpha)-dehydrogenase from Rhodococcus jostii RHA1 in complex with 4-androstene-3,17- dione | | Descriptor: | 3-KETOSTEROID-DELTA4-5ALPHA-DEHYDROGENASE, 4-ANDROSTENE-3-17-DIONE, CHLORIDE ION, ... | | Authors: | van Oosterwijk, N, Knol, J, Dijkhuizen, L, van der Geize, R, Dijkstra, B.W. | | Deposit date: | 2012-05-03 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Catalytic Mechanism of 3-Ketosteroid-{Delta}4-(5Alpha)-Dehydrogenase from Rhodococcus Jostii Rha1 Genome.
J.Biol.Chem., 287, 2012
|
|
4C2L
 
 | | Crystal structure of endo-xylogalacturonan hydrolase from Aspergillus tubingensis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-XYLOGALACTURONAN HYDROLASE A, ... | | Authors: | Rozeboom, H.J, Beldman, G, Schols, H.A, Dijkstra, B.W. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Endo-Xylogalacturonan Hydrolase from Aspergillus Tubingensis.
FEBS J., 280, 2013
|
|
4C5R
 
 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | (3S)-3-amino-2,2-difluoro-3-phenylpropanoic acid, GLYCEROL, PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
4C5S
 
 | | Structural Investigations into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus chinensis | | Descriptor: | (3S)-3-amino-2,2-difluoro-3-phenylpropanoic acid, PHENYLALANINE AMMONIA-LYASE | | Authors: | Wybenga, G.G, Szymanski, W, Wu, B, Feringa, B.L, Janssen, D.B, Dijkstra, B.W. | | Deposit date: | 2013-09-16 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Investigations Into the Stereochemistry and Activity of a Phenylalanine-2,3-Aminomutase from Taxus Chinensis.
Biochemistry, 53, 2014
|
|
