5D5A
 
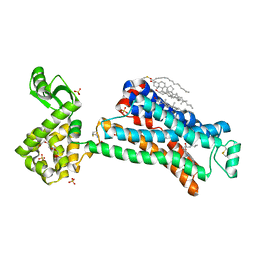 | | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Liu, X, Kobilka, B, Kay Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4826 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4DF8
 
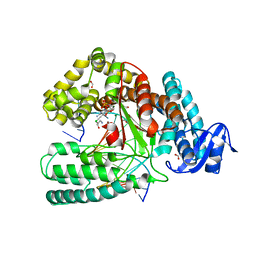 | | Crystal structure of the large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with aminopentinyl-7-deaza-2-dATP | | Descriptor: | 1,2-ETHANEDIOL, 5-(5-aminopent-1-yn-1-yl)-7-{2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, CHLORIDE ION, ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
4DF4
 
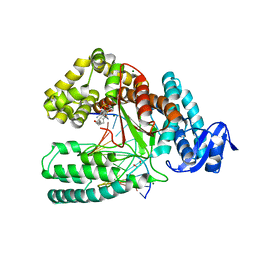 | | Crystal structure of the large fragment of DNA Polymerase I from Thermus aquaticus in a closed ternary complex with 7-(N-(10-hydroxydecanoyl)-aminopentinyl)-7-deaza-2 -dATP | | Descriptor: | 1,2-ETHANEDIOL, 7-{2-deoxy-5-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-erythro-pentofuranosyl}-5-{5-[(10-hydroxydecanoyl)amino]pent-1-yn-1-yl}-7H-pyrrolo[2,3-d]pyrimidin-4-amine, DNA (5'-D(*AP*AP*AP*TP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Bergen, K, Steck, A, Struett, S, Baccaro, A, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of KlenTaq DNA Polymerase Caught While Incorporating C5-Modified Pyrimidine and C7-Modified 7-Deazapurine Nucleoside Triphosphates.
J.Am.Chem.Soc., 134, 2012
|
|
1C1Z
 
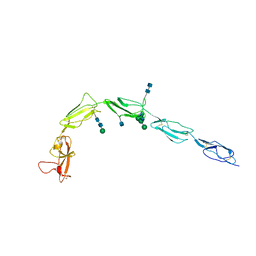 | | CRYSTAL STRUCTURE OF HUMAN BETA-2-GLYCOPROTEIN-I (APOLIPOPROTEIN-H) | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA2-GLYCOPROTEIN-I, ... | | Authors: | Schwarzenbacher, R, Zeth, K, Diederichs, K, Gries, A, Kostner, G.M, Laggner, P, Prassl, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-11-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome.
EMBO J., 18, 1999
|
|
1CS6
 
 | | N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN | | Descriptor: | AXONIN-1, GLYCEROL | | Authors: | Freigang, J, Proba, K, Diederichs, K, Sonderegger, P, Welte, W. | | Deposit date: | 1999-08-17 | | Release date: | 2000-05-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion.
Cell(Cambridge,Mass.), 101, 2000
|
|
3T3F
 
 | | Ternary Structure of the large fragment of Taq DNA polymerase bound to an abasic site and dNITP | | Descriptor: | 1-{2-DEOXY-5-O-[(R)-HYDROXY{[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}PHOSPHORYL]-BETA-D-ERYTHRO-PENTOFURANOSYL}-5-NITRO -1H-INDOLE, 5'-D(*AP*AP*AP*(3DR)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3', ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amino acid templating mechanisms in selection of nucleotides opposite abasic sites by a family a DNA polymerase.
J.Biol.Chem., 287, 2012
|
|
4NSI
 
 | | Carboplatin binding to HEWL in 20% propanol, 20% PEG 4000 at pH5.6 | | Descriptor: | CITRATE ANION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4NSG
 
 | | Carboplatin binding to HEWL in NaBr crystallisation conditions studied at an X-ray wavelength of 1.5418A | | Descriptor: | ACETATE ION, BROMIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4NSJ
 
 | | Carboplatin binding to HEWL in 2M NH4formate, 0.1M HEPES at pH 7.5 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4NSH
 
 | | Carboplatin binding to HEWL in 0.2M NH4SO4, 0.1M NaAc in 25% PEG 4000 at pH 4.6 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Tanley, S.W.M, Diederichs, K, Kroon-Batenburg, L.M.J, Levy, C, Schreurs, A.M.M, Helliwell, J.R. | | Deposit date: | 2013-11-28 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Carboplatin binding to histidine.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4BWM
 
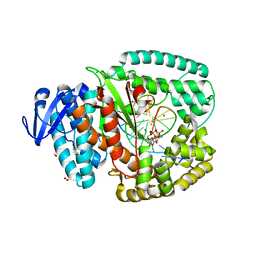 | | KlenTaq mutant in complex with a RNA/DNA hybrid | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOCP)-3', 5'-R(*AP*AP*AP*GP*GP*GP*CP*GP*CP*CP*GP*UP*GP*GP*UP*C)-3', ... | | Authors: | Blatter, N, Bergen, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-07-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure and Function of an RNA-Reading Thermostable DNA Polymerase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
4BWJ
 
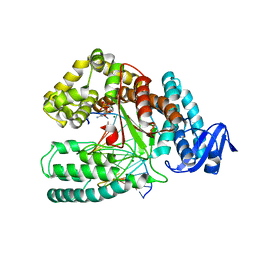 | | KlenTaq mutant in complex with DNA and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, 5'-D(*AP*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOCP)-3', ... | | Authors: | Blatter, N, Bergen, K, Welte, W, Diederichs, K, Marx, A. | | Deposit date: | 2013-07-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure and Function of an RNA-Reading Thermostable DNA Polymerase.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1DUW
 
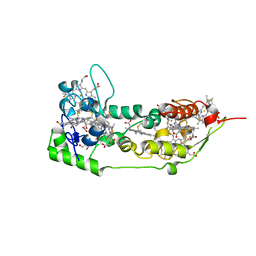 | | STRUCTURE OF NONAHEME CYTOCHROME C | | Descriptor: | GLYCEROL, HEME C, NONAHEME CYTOCHROME C | | Authors: | Umhau, S, Fritz, G, Diederichs, K, Breed, J, Kroneck, P.M, Welte, W. | | Deposit date: | 2000-01-19 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Three-dimensional structure of the nonaheme cytochrome c from Desulfovibrio desulfuricans Essex in the Fe(III) state at 1.89 A resolution.
Biochemistry, 40, 2001
|
|
1FCP
 
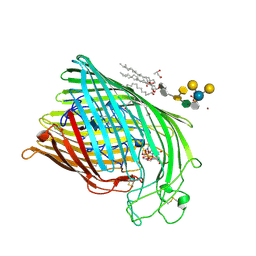 | | FERRIC HYDROXAMATE UPTAKE RECEPTOR (FHUA) FROM E.COLI IN COMPLEX WITH BOUND FERRICHROME-IRON | | Descriptor: | 2-TRIDECANOYLOXY-PENTADECANOIC ACID, 3-OXO-PENTADECANOIC ACID, ACETOACETIC ACID, ... | | Authors: | Hofmann, E, Ferguson, A.D, Diederichs, K, Welte, W. | | Deposit date: | 1998-10-14 | | Release date: | 1999-01-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Siderophore-mediated iron transport: crystal structure of FhuA with bound lipopolysaccharide.
Science, 282, 1998
|
|
1EU8
 
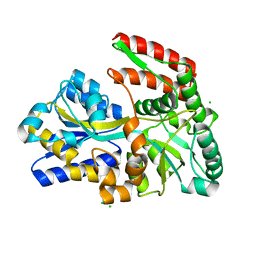 | | STRUCTURE OF TREHALOSE MALTOSE BINDING PROTEIN FROM THERMOCOCCUS LITORALIS | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, TREHALOSE/MALTOSE BINDING PROTEIN, ... | | Authors: | Diez, J, Diederichs, K, Greller, G, Horlacher, R, Boos, W, Welte, W. | | Deposit date: | 2000-04-14 | | Release date: | 2001-03-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic Archaeon Thermococcus litoralis at 1.85 A.
J.Mol.Biol., 305, 2001
|
|
1FUE
 
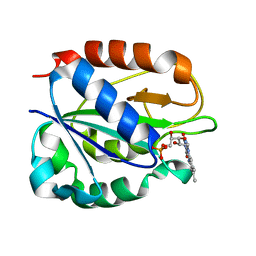 | | FLAVODOXIN FROM HELICOBACTER PYLORI | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Freigang, J, Diederichs, K, Schaefer, K.P, Welte, W, Paul, R. | | Deposit date: | 2000-09-15 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of oxidized flavodoxin, an essential protein in Helicobacter pylori.
Protein Sci., 11, 2002
|
|
1T92
 
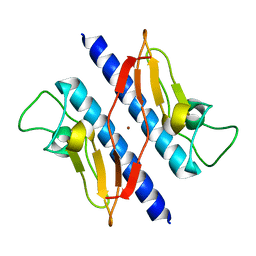 | | Crystal structure of N-terminal truncated pseudopilin PulG | | Descriptor: | General secretion pathway protein G, ZINC ION | | Authors: | Koehler, R, Schaefer, K, Mueller, S, Vignon, G, Diederichs, K, Philippsen, A, Ringler, P, Pugsley, A.P, Engel, A, Welte, W. | | Deposit date: | 2004-05-14 | | Release date: | 2004-10-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and assembly of the pseudopilin PulG.
Mol.Microbiol., 54, 2004
|
|
1URS
 
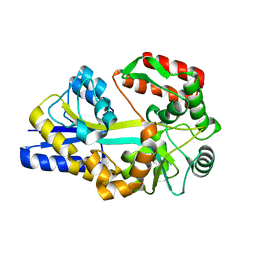 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-11-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins
J.Mol.Biol., 335, 2004
|
|
1URD
 
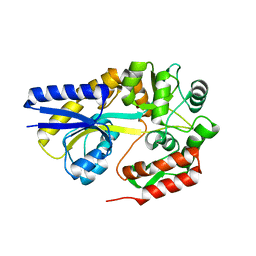 | | X-ray structures of the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius provide insight into acid stability of proteins | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1URG
 
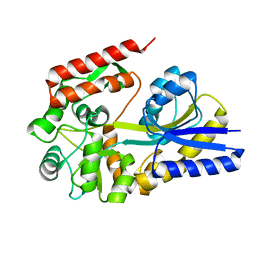 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
4ELU
 
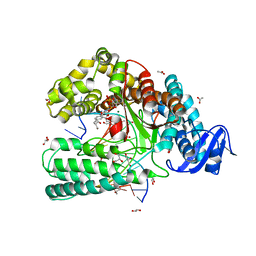 | | Snapshot of the large fragment of DNA polymerase I from Thermus Aquaticus processing modified pyrimidines | | Descriptor: | 2'-deoxy-5-[(4-ethynylphenyl)ethynyl]cytidine 5'-(tetrahydrogen triphosphate), ACETATE ION, DNA (5'-D(*AP*AP*AP*GP*CP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), ... | | Authors: | Marx, A, Diederichs, K, Obeid, S. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interactions of non-polar and "Click-able" nucleotides in the confines of a DNA polymerase active site.
Chem.Commun.(Camb.), 48, 2012
|
|
4X5U
 
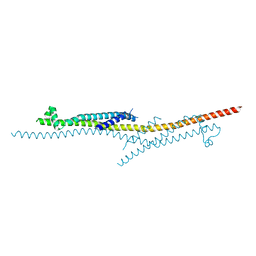 | | X-ray crystal structure of CagL at pH 4.2 | | Descriptor: | Cag pathogenicity island protein (Cag18) | | Authors: | Sundberg, E.J, Bonsor, D.A, Diederichs, K. | | Deposit date: | 2014-12-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Integrin Engagement by the Helical RGD Motif of the Helicobacter pylori CagL Protein Is Regulated by pH-induced Displacement of a Neighboring Helix.
J.Biol.Chem., 290, 2015
|
|
4XJB
 
 | | X-ray structure of Lysozyme1 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJG
 
 | | X-ray structure of Lysozyme B2 | | Descriptor: | BROMIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XJI
 
 | | X-ray structure of LysozymeS2 | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Huang, C.Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-01-08 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
