9F7K
 
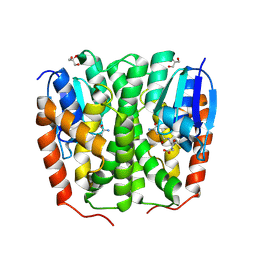 | | Glutathione transferase epsilon 1 from Drosophila melanogaster in complex with glutathione | | Descriptor: | GH14654p, GLUTATHIONE, GLYCEROL, ... | | Authors: | Didierjean, C, Schwartz, M, Neiers, F. | | Deposit date: | 2024-05-03 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Thermodynamic Insights into Dimerization Interfaces of Drosophila Glutathione Transferases.
Biomolecules, 14, 2024
|
|
7PWE
 
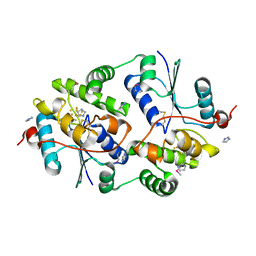 | |
8AI9
 
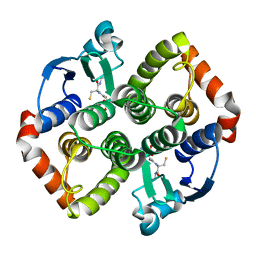 | |
8AIB
 
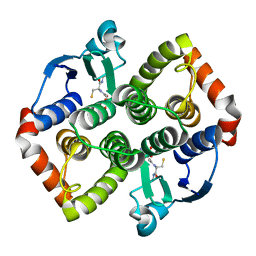 | |
8AI8
 
 | |
4LMW
 
 | |
7PKA
 
 | | Synechocystis sp. PCC6803 glutathione transferase Chi 1, GSOH bound | | Descriptor: | Glutathione S-transferase, POTASSIUM ION, S-Hydroxy-Glutathione | | Authors: | Didierjean, C, Mocchetti, E, Hecker, A, Favier, F. | | Deposit date: | 2021-08-25 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure and functions of glutathione transferase Chi 1 from cyanobacterium Synechocystis sp. PCC 6803
To Be Published
|
|
2DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NADP+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-19 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
4LMV
 
 | |
3FZ9
 
 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
3FZA
 
 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione and beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
7ZVP
 
 | |
7ZZN
 
 | |
8A0I
 
 | |
8A0O
 
 | |
8A08
 
 | |
8A0R
 
 | |
8A0Q
 
 | |
8A0P
 
 | | Crystal structure of poplar glutathione transferase U20 in complex with morin | | Descriptor: | 2-[2,4-bis(oxidanyl)phenyl]-3,5,7-tris(oxidanyl)chromen-4-one, CHLORIDE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-30 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
7ZS3
 
 | | Crystal structure of poplar glutathione transferase U19 | | Descriptor: | ACETATE ION, Glutathione transferase | | Authors: | Didierjean, C, Favier, F. | | Deposit date: | 2022-05-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Biochemical and Structural Insights on the Poplar Tau Glutathione Transferase GSTU19 and 20 Paralogs Binding Flavonoids.
Front Mol Biosci, 9, 2022
|
|
5EY6
 
 | |
5F06
 
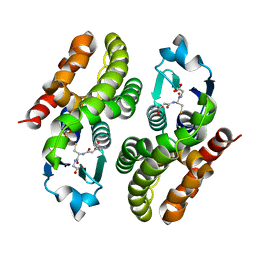 | | Crystal structure of glutathione transferase F7 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, Glutathione S-transferase family protein, SULFATE ION | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
1DBV
 
 | | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE MUTANT WITH ASP 32 REPLACED BY GLY, LEU 187 REPLACED BY ALA, AND PRO 188 REPLACED BY SER COMPLEXED WITH NAD+ | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Rahuel-Clermont, S, Vitoux, B, Dideberg, O, Branlant, G, Aubry, A. | | Deposit date: | 1996-12-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A crystallographic comparison between mutated glyceraldehyde-3-phosphate dehydrogenases from Bacillus stearothermophilus complexed with either NAD+ or NADP+.
J.Mol.Biol., 268, 1997
|
|
5F07
 
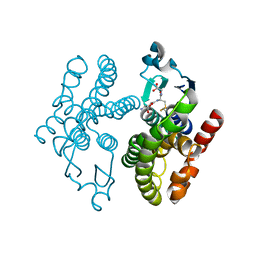 | | Crystal structure of glutathione transferase F8 from Populus trichocarpa | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase family protein | | Authors: | Didierjean, C, Rouhier, N, Pegeot, H, Gense, F. | | Deposit date: | 2015-11-27 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural plasticity among glutathione transferase Phi members: natural combination of catalytic residues confers dual biochemical activities.
FEBS J., 284, 2017
|
|
3PPU
 
 | |
