8IBX
 
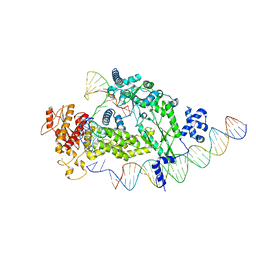 | | Structure of R2 with 3'UTR and DNA in unwinding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBW
 
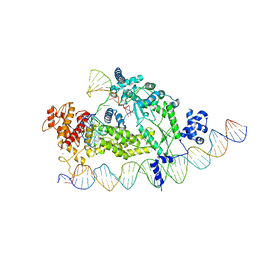 | | Structure of R2 with 3'UTR and DNA in binding state | | Descriptor: | 3'UTR, DNA (60-MER), Reverse transcriptase-like protein, ... | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBZ
 
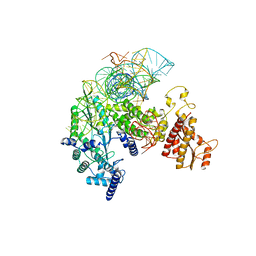 | | Structure of R2 with 5'ORF and 3'UTR | | Descriptor: | 5ORF-linker-3UTR, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
8IBY
 
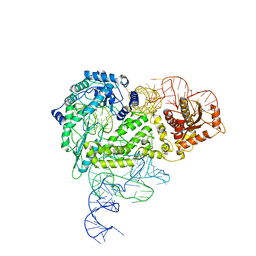 | | Structure of R2 with 5'ORF | | Descriptor: | 5'ORF RNA, Reverse transcriptase-like protein, ZINC ION | | Authors: | Deng, P, Tan, S, Wang, J, Liu, J.J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural RNA components supervise the sequential DNA cleavage in R2 retrotransposon.
Cell, 186, 2023
|
|
4XEY
 
 | |
1QFD
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-AMYLASE INHIBITOR (AAI) | | Descriptor: | PROTEIN (ALPHA-AMYLASE INHIBITOR) | | Authors: | Lu, S, Deng, P, Liu, X, Luo, J, Han, R, Gu, X, Liang, S, Wang, X, Feng, L, Lozanov, V, Patthy, A, Pongor, S. | | Deposit date: | 1999-04-08 | | Release date: | 1999-07-16 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the major alpha-amylase inhibitor of the crop plant amaranth.
J.Biol.Chem., 274, 1999
|
|
1V07
 
 | | CRYSTAL STRUCTURE OF ThrE11Val mutant of THE NERVE TISSUE MINI-HEMOGLOBIN FROM THE NEMERTEAN WORM CEREBRATULUS LACTEUS | | Descriptor: | NEURAL HEMOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Pesce, A, Nardini, M, Ascenzi, P, Geuens, E, Dewilde, S, Moens, L, Bolognesi, M, Riggs, A, Hale, A, Deng, P, Nienhaus, G.U, Olson, J.S, Nienhaus, K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thr-E11 Regulates O2 Affinity in Cerebratulus Lacteus Mini-Hemoglobin
J.Biol.Chem., 279, 2004
|
|
4LGB
 
 | | ABA-mimicking ligand N-(1-METHYL-2-OXO-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-(4-METHYLPHENYL)METHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(1-methyl-2-oxo-1,2,3,4-tetrahydroquinolin-6-yl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LG5
 
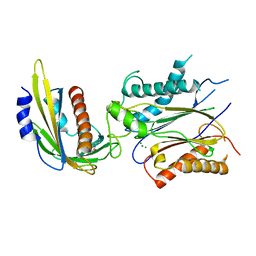 | | ABA-mimicking ligand QUINABACTIN in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, Protein phosphatase 2C 16, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
4LGA
 
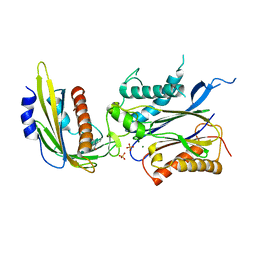 | | ABA-mimicking ligand N-(2-OXO-1-PROPYL-1,2,3,4-TETRAHYDROQUINOLIN-6-YL)-1-PHENYLMETHANESULFONAMIDE in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | Abscisic acid receptor PYL2, MAGNESIUM ION, N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)-1-phenylmethanesulfonamide, ... | | Authors: | Zhou, X.E, Gao, M, Liu, X, Zhang, Y, Xue, X, Melcher, K, Gao, P, Wang, F, Zeng, L, Zhao, Y, Zhao, Y, Deng, P, Zhong, D, Zhu, J.-K, Xu, Y, Xu, H.E. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants.
Cell Res., 23, 2013
|
|
6AF0
 
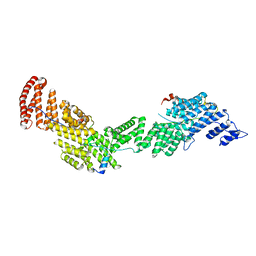 | | Structure of Ctr9, Paf1 and Cdc73 ternary complex from Myceliophthora thermophila | | Descriptor: | Cdc73 protein, Ctr9 protein, Paf1 protein | | Authors: | Wang, Z, Deng, P, Zhou, Y. | | Deposit date: | 2018-08-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Transcriptional elongation factor Paf1 core complex adopts a spirally wrapped solenoidal topology.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7V59
 
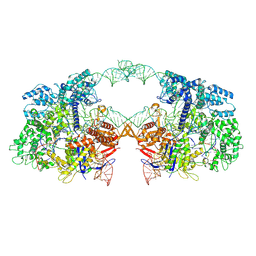 | | Cryo-EM structure of spyCas9-sgRNA-DNA dimer | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (49-MER), RNA (115-MER) | | Authors: | Liu, J, Deng, P. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (5.26 Å) | | Cite: | Nonspecific interactions between SpCas9 and dsDNA sites located downstream of the PAM mediate facilitated diffusion to accelerate target search.
Chem Sci, 12, 2021
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2021-03-03 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
