7JRQ
 
 | | Crystallographically Characterized De Novo Designed Mn-Diphenylporphyrin Binding Protein | | 分子名称: | CHLORIDE ION, GLYCEROL, MPP1, ... | | 著者 | Mann, S.I, DeGrado, W.F. | | 登録日 | 2020-08-12 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | De Novo Design, Solution Characterization, and Crystallographic Structure of an Abiological Mn-Porphyrin-Binding Protein Capable of Stabilizing a Mn(V) Species.
J.Am.Chem.Soc., 143, 2021
|
|
8TNB
 
 | |
8TNC
 
 | |
8TN1
 
 | |
8TN6
 
 | |
8TND
 
 | |
7UDV
 
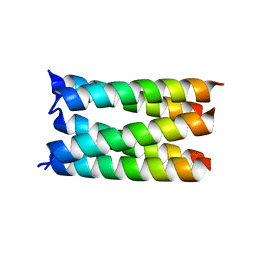 | |
7UDZ
 
 | | Designed pentameric proton channel LQLL | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, De novo designed pentameric proton channel LQLL | | 著者 | Kratochvil, H.T, Thomaston, J.L, Mravic, M, Nicoludis, J.M, Liu, L, DeGrado, W.F. | | 登録日 | 2022-03-20 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.48 Å) | | 主引用文献 | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
7UDY
 
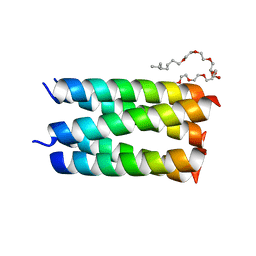 | | Designed pentameric channel QLLL | | 分子名称: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, Designed channel QLLL | | 著者 | Kratochvil, H.T, Thomaston, J.L, Liu, L, DeGrado, W.F. | | 登録日 | 2022-03-20 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Transient water wires mediate selective proton transport in designed channel proteins.
Nat.Chem., 15, 2023
|
|
6YB1
 
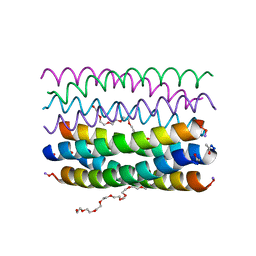 | | Crystal structure of an antiparallel octameric transmembrane coiled coil K2-CCTM-VbIc | | 分子名称: | GLYCINE, K2-CCTM-VbIc, NONAETHYLENE GLYCOL | | 著者 | Kratochvil, H.T, Liu, L, Scott, A.J, Woolfson, D.N, DeGrado, W.F. | | 登録日 | 2020-03-15 | | 公開日 | 2021-04-07 | | 最終更新日 | 2021-07-14 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Constructing ion channels from water-soluble alpha-helical barrels.
Nat.Chem., 13, 2021
|
|
1QP6
 
 | |
3V86
 
 | |
3UUG
 
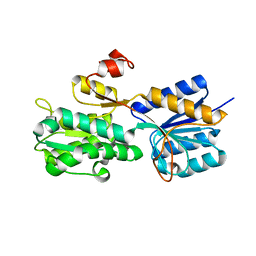 | | Crystal structure of the periplasmic sugar binding protein ChvE | | 分子名称: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-glucopyranuronic acid | | 著者 | Hu, X, Zhao, J, Binns, A, Degrado, W. | | 登録日 | 2011-11-28 | | 公開日 | 2012-11-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3URM
 
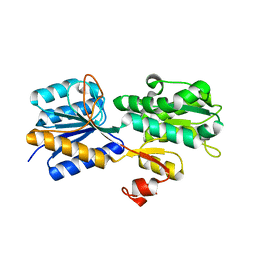 | | Crystal structure of the periplasmic sugar binding protein ChvE | | 分子名称: | Multiple sugar-binding periplasmic receptor ChvE, beta-D-galactopyranose | | 著者 | Hu, X, Zhao, J, Binns, A, Degrado, W. | | 登録日 | 2011-11-22 | | 公開日 | 2012-11-28 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.801 Å) | | 主引用文献 | Agrobacterium tumefaciens recognizes its host environment using ChvE to bind diverse plant sugars as virulence signals.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6O3N
 
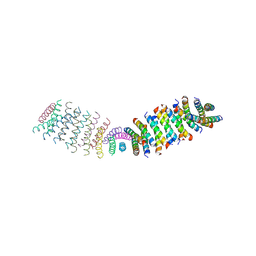 | |
1YOD
 
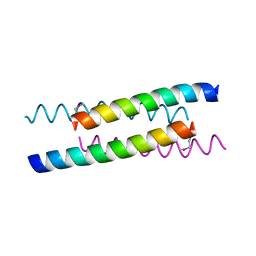 | |
1Y47
 
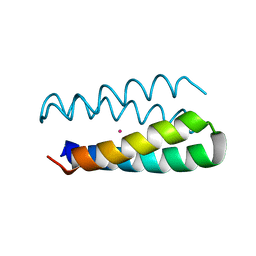 | | Structural studies of designed alpha-helical hairpins | | 分子名称: | CADMIUM ION, dueferri (DF2) | | 著者 | Lahr, S.J, Engel, D.E, Stayrook, S.E, Maglio, O, North, B, Geremia, S, Lombardi, A, DeGrado, W.F. | | 登録日 | 2004-11-30 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Analysis and design of turns in alpha-helical hairpins
J.Mol.Biol., 346, 2005
|
|
6OUG
 
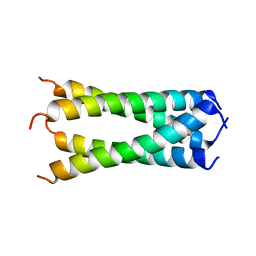 | | Structure of drug-resistant V27A mutant of the influenza M2 proton channel bound to spiroadamantyl amine inhibitor, TM + cytosolic helix construct | | 分子名称: | (1r,1'S,3'S,5'S,7'S)-spiro[cyclohexane-1,2'-tricyclo[3.3.1.1~3,7~]decan]-4-amine, Matrix protein 2 | | 著者 | Thomaston, J.L, Liu, L, DeGrado, W.F. | | 登録日 | 2019-05-04 | | 公開日 | 2020-01-15 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | X-ray Crystal Structures of the Influenza M2 Proton Channel Drug-Resistant V27A Mutant Bound to a Spiro-Adamantyl Amine Inhibitor Reveal the Mechanism of Adamantane Resistance.
Biochemistry, 59, 2020
|
|
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | 分子名称: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | 著者 | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | 登録日 | 2017-01-09 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2KZ2
 
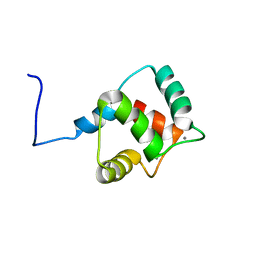 | | Calmodulin, C-terminal domain, F92E mutant | | 分子名称: | CALCIUM ION, Calmodulin | | 著者 | Korendovych, I, Kulp, D, Wu, Y, Cheng, H, Roder, H, DeGrado, W. | | 登録日 | 2010-06-10 | | 公開日 | 2011-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Design of a switchable eliminase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6Z0L
 
 | | Het-N2 - De novo designed three-helix heterodimer with Cysteine at the N2 position of the alpha-helix | | 分子名称: | CADMIUM ION, Cys-N2 Strand, Positive Strand, ... | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | 登録日 | 2020-05-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
6Z0M
 
 | | Het-Ncap - De novo designed three-helix heterodimer with Cysteine at the Ncap position of the alpha-helix | | 分子名称: | Cys-Ncap strand, Positive Strand, SULFATE ION, ... | | 著者 | McEwen, A.G, Poussin-Courmontagne, P, Naudin, E.A, DeGrado, W.F, Torbeev, V. | | 登録日 | 2020-05-09 | | 公開日 | 2021-03-17 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Acyl Transfer Catalytic Activity in De Novo Designed Protein with N-Terminus of alpha-Helix As Oxyanion-Binding Site.
J.Am.Chem.Soc., 143, 2021
|
|
1AL1
 
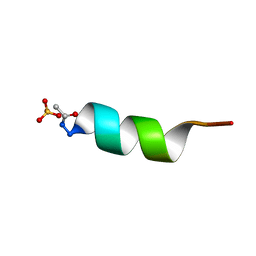 | | CRYSTAL STRUCTURE OF ALPHA1: IMPLICATIONS FOR PROTEIN DESIGN | | 分子名称: | ALPHA HELIX PEPTIDE: ELLKKLLEELKG, SULFATE ION | | 著者 | Hill, C.P, Anderson, D.H, Wesson, L, Degrado, W.F, Eisenberg, D. | | 登録日 | 1990-07-02 | | 公開日 | 1991-10-15 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structure of alpha 1: implications for protein design.
Science, 249, 1990
|
|
4JG0
 
 | | Structure of phosphoserine/threonine (pSTAb) scaffold bound to pSer peptide | | 分子名称: | Fab heavy chain, Fab light chain, PROPANOIC ACID, ... | | 著者 | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | 登録日 | 2013-02-28 | | 公開日 | 2013-08-28 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
4JFX
 
 | | Structure of phosphotyrosine (pTyr) scaffold bound to pTyr peptide | | 分子名称: | Fab heavy chain, Fab light chain, Phosphopeptide, ... | | 著者 | Koerber, J.T, Thomsen, N.D, Hannigan, B.T, Degrado, W.F, Wells, J.A. | | 登録日 | 2013-02-28 | | 公開日 | 2013-09-25 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Nature-inspired design of motif-specific antibody scaffolds.
Nat.Biotechnol., 31, 2013
|
|
