2ADT
 
 | | NMR structure of a 30 kDa GAAA tetraloop-receptor complex. | | Descriptor: | 43-MER | | Authors: | Davis, J.H, Tonelli, M, Scott, L.G, Jaeger, L, Williamson, J.R, Butcher, S.E. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | RNA Helical Packing in Solution: NMR Structure of a 30 kDa GAAA Tetraloop-Receptor Complex
J.Mol.Biol., 351, 2005
|
|
3EE1
 
 | |
1OMC
 
 | | SOLUTION STRUCTURE OF OMEGA-CONOTOXIN GVIA USING 2-D NMR SPECTROSCOPY AND RELAXATION MATRIX ANALYSIS | | Descriptor: | OMEGA-CONOTOXIN GVIA | | Authors: | Davis, J.H, Bradley, E.K, Miljanich, G.P, Nadasdi, L, Ramachandran, J, Basus, V.J. | | Deposit date: | 1993-04-28 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega-conotoxin GVIA using 2-D NMR spectroscopy and relaxation matrix analysis.
Biochemistry, 32, 1993
|
|
2I7Z
 
 | |
2I7E
 
 | |
7QEL
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247 | | Descriptor: | (2~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)spiro[2.3]hexane-2-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2021-12-03 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011247
To Be Published
|
|
6TWO
 
 | | Binding domain of BoNT/A6 | | Descriptor: | Bont/A1, CHLORIDE ION | | Authors: | Davies, J.R, Britton, A, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
6TWP
 
 | | Binding domain of BoNT/A5 | | Descriptor: | Botulinum neurotoxin A5, CHLORIDE ION | | Authors: | Davies, J.R, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
7AZ0
 
 | |
8THJ
 
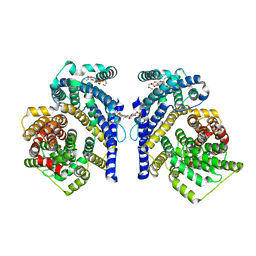 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, PHOSPHATIDYLETHANOLAMINE, SODIUM ION, ... | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
8THI
 
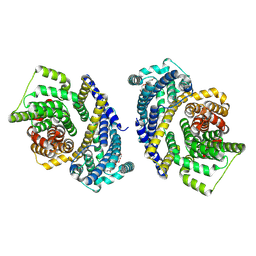 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, SODIUM ION, Sialic acid TRAP transporter permease protein SiaT | | Authors: | Davies, J.S, Currie, M.C, Dobson, R.C.J, North, R.A. | | Deposit date: | 2023-07-16 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural and biophysical analysis of a Haemophilus influenzae tripartite ATP-independent periplasmic (TRAP) transporter.
Elife, 12, 2024
|
|
7T3E
 
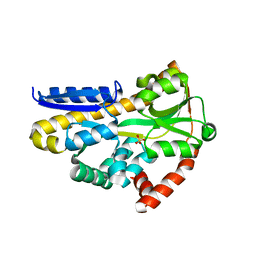 | | Structure of the sialic acid bound Tripartite ATP-independent Periplasmic (TRAP) periplasmic component SiaP from Photobacterium profundum | | Descriptor: | N-acetyl-beta-neuraminic acid, SULFATE ION, TRAP-type C4-dicarboxylate transport system, ... | | Authors: | Davies, J.S, Currie, M.J, North, R.A, Dobson, R.C.J. | | Deposit date: | 2021-12-07 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
8B01
 
 | | Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc | | Descriptor: | DECANE, DOCOSANE, HEXANE, ... | | Authors: | Davies, J.S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2022-09-06 | | Release date: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structure and mechanism of a tripartite ATP-independent periplasmic TRAP transporter.
Nat Commun, 14, 2023
|
|
6ZVN
 
 | |
6ZVM
 
 | | Botulinum neurotoxin B2 binding domain in complex with GD1a | | Descriptor: | GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Neurotoxin | | Authors: | Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2020-07-25 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Characterization of Botulinum Neurotoxin Subtype B2 Binding to Its Receptors.
Toxins, 12, 2020
|
|
7Z5R
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035 | | Descriptor: | (7~{R})-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)bicyclo[4.2.0]octa-1,3,5-triene-7-carboxamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-09 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012035
To Be Published
|
|
7ZG3
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228 | | Descriptor: | N-glycosylase/DNA lyase, NICKEL (II) ION, ~{N}-[(1~{S})-1,2,2-trimethylcyclopropyl]pyrrolo[1,2-c]pyrimidine-3-carboxamide | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-04-01 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH011228
To Be Published
|
|
6F0P
 
 | | Botulinum neurotoxin A4 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Davies, J.R, Rees, J, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
6F0O
 
 | | Botulinum neurotoxin A3 Hc domain | | Descriptor: | 1,2-ETHANEDIOL, Bontoxilysin A, PENTAETHYLENE GLYCOL, ... | | Authors: | Davies, J.R, Liu, S.M, Acharya, K.R. | | Deposit date: | 2017-11-20 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution crystal structures of Clostridium botulinum neurotoxin A3 and A4 binding domains.
J. Struct. Biol., 202, 2018
|
|
7ZC7
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941 | | Descriptor: | 2-[4-(3,5-dimethylpyrazol-1-yl)phenyl]-~{N}-(4,5,6,7-tetrahydro-1,2-benzoxazol-3-yl)ethanamide, N-glycosylase/DNA lyase, NICKEL (II) ION | | Authors: | Davies, J.R, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-03-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH012941
To Be Published
|
|
5MK8
 
 | |
3CF2
 
 | | Structure of P97/vcp in complex with ADP/AMP-PNP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
3CF1
 
 | | Structure of P97/vcp in complex with ADP/ADP.alfx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Transitional endoplasmic reticulum ATPase | | Authors: | Davies, J.M, Delabarre, B, Brunger, A.T, Weis, W.I. | | Deposit date: | 2008-03-01 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change.
Structure, 16, 2008
|
|
5MK6
 
 | |
5MK7
 
 | |
