5L7U
 
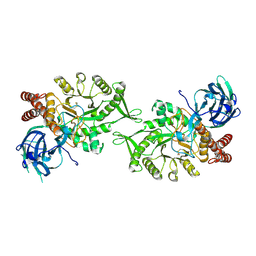 | | Crystal structure of BvGH123 with bound GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, CHLORIDE ION, Glycoside hydrolase | | Authors: | Roth, C, Petricevic, M, John, A, Goddard-Borger, E.D, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-06-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic insights into a Bacteroides vulgatus retaining N-acetyl-beta-galactosaminidase that uses neighbouring group participation.
Chem. Commun. (Camb.), 52, 2016
|
|
4D1J
 
 | | The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicas in complex with 1-Deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Larsbrink, J, Thompson, A.J, Lundqvist, M, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complex Gene Locus Enables Xyloglucan Utilization in the Model Saprophyte Cellvibrio Japonicus.
Mol.Microbiol., 94, 2014
|
|
4D4B
 
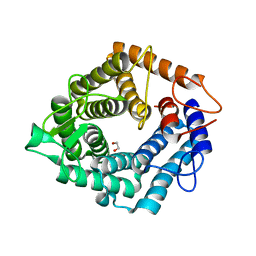 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with MSMSMe | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE, alpha-D-mannopyranose-(1-6)-methyl 1,6-dithio-alpha-D-mannopyranoside | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4C7F
 
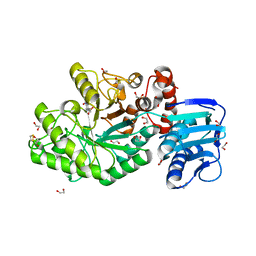 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyen Thi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
4C7G
 
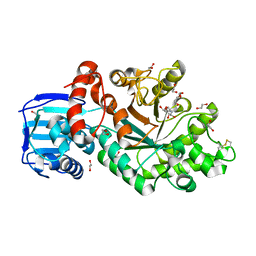 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
4BMZ
 
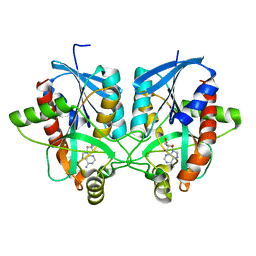 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4D4C
 
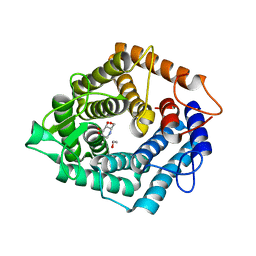 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManDMJ | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D4D
 
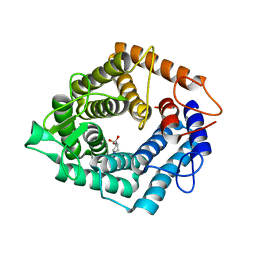 | | The catalytic domain, BcGH76, of Bacillus circulans Aman6 in complex with 1,6-ManIFG | | Descriptor: | 1,2-ETHANEDIOL, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ALPHA-1,6-MANNANASE, ... | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4D1I
 
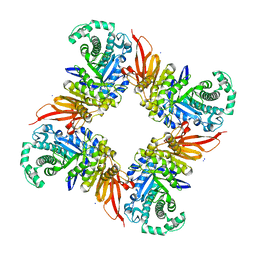 | | The structure of the GH35 beta-galactosidase Bgl35A from Cellvibrio japonicus | | Descriptor: | ACETATE ION, BETA-GALACTOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Thompson, A.J, Lundqvist, M, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Complex Gene Locus Enables Xyloglucan Utilization in the Model Saprophyte Cellvibrio Japonicus.
Mol.Microbiol., 94, 2014
|
|
4BMH
 
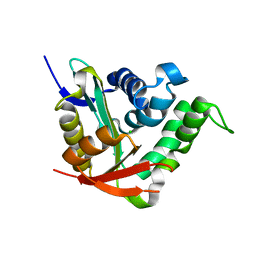 | | Crystal structure of SsHAT | | Descriptor: | ACETYLTRANSFERASE, CHLORIDE ION | | Authors: | He, Y, Turkenburg, J.P, Davies, G.J. | | Deposit date: | 2013-05-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of a Streptomyces Sviceus Gnat Acetyltransferase with Similarity to the C-Terminal Domain of the Human Gh84 O-Glcnacase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C7D
 
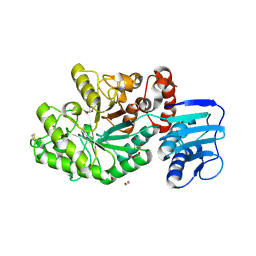 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
4C1R
 
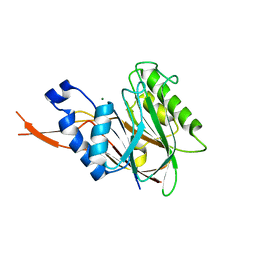 | | Bacteroides thetaiotaomicron VPI-5482 mannosyl-6-phosphatase Bt3783 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MANNOSYL-6-PHOSPHATASE | | Authors: | Cuskin, F, Lowe, E.C, Zhu, Y, Temple, M, Thompson, A.J, Cartmell, A, Piens, K, Bracke, D, Vervecken, W, Munoz-Munoz, J.L, Suits, M.D.L, Boraston, A.B, Williams, S.J, Davies, G.J, Abbott, W.D, Martens, E.C, Gilbert, H.J. | | Deposit date: | 2013-08-13 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Human Gut Bacteroidetes Can Utilize Yeast Mannan Through a Selfish Mechanism.
Nature, 517, 2015
|
|
4D4A
 
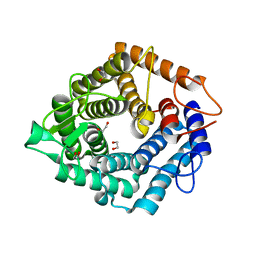 | | Structure of the catalytic domain (BcGH76) of the Bacillus circulans GH76 alpha mannanase, Aman6. | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-1,6-MANNANASE | | Authors: | Thompson, A.J, Speciale, G, Iglesias-Fernandez, J, Hakki, Z, Belz, T, Cartmell, A, Spears, R.J, Stepper, J, Gilbert, H.J, Rovira, C, Williams, S.J, Davies, G.J. | | Deposit date: | 2014-10-27 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evidence for a Boat Conformation at the Transition State of Gh76 Alpha-1,6-Mannanases- Key Enzymes in Bacterial and Fungal Mannoprotein Metabolism
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4BN0
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BMY
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | MTA/SAH NUCLEOSIDASE, SULFATE ION | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4UTT
 
 | | Structural characterisation of NanE, ManNac6P C2 epimerase, from Clostridium perfingens | | Descriptor: | ACETATE ION, CHLORIDE ION, PUTATIVE N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway.
J.Biol.Chem., 289, 2014
|
|
6YP1
 
 | | HiCel7B unliganded | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, Endoglucanase 1, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Glycosylated cyclophellitol-derived activity-based probes and inhibitors for cellulases.
Rsc Chem Biol, 1, 2020
|
|
6YJS
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJQ
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A,Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJT
 
 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YQH
 
 | | GH146 beta-L-arabinofuranosidase bound to covalent inhibitor | | Descriptor: | (1~{S},2~{S},3~{S},4~{S})-4-(hydroxymethyl)cyclopentane-1,2,3-triol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acetyl-CoA carboxylase, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-17 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Cysteine Nucleophiles in Glycosidase Catalysis: Application of a Covalent beta-l-Arabinofuranosidase Inhibitor.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YJV
 
 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP-2-deoxy-2-fluoroglucose and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YOZ
 
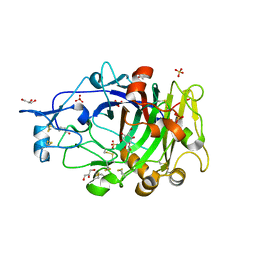 | | HiCel7B labelled with b-1,4-glucosyl cyclophellitol | | Descriptor: | (1R,2S,3S,4S,5R,6R)-6-(HYDROXYMETHYL)CYCLOHEXANE-1,2,3,4,5-PENTOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETAMIDE, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-04-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Glycosylated cyclophellitol-derived activity-based probes and inhibitors for cellulases.
Rsc Chem Biol, 1, 2020
|
|
6YJU
 
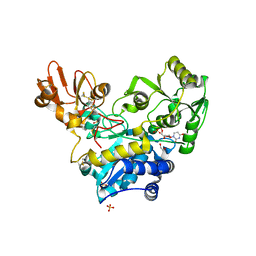 | | Crystal structure of MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain with a Lys329-Ile345 loop truncation, in complex with UDP and biantennary pentasaccharide M592 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
6YJR
 
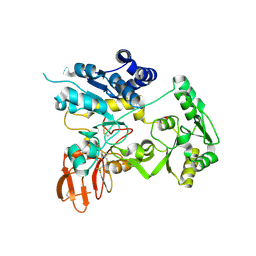 | | Crystal structure of unliganded MGAT5 (alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase V) luminal domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase A, ... | | Authors: | Wu, L, Darby, J.F, Gilio, A.K, Davies, G.J. | | Deposit date: | 2020-04-04 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Substrate Engagement and Catalytic Mechanisms of N-Acetylglucosaminyltransferase V
Acs Catalysis, 2020
|
|
