1W2P
 
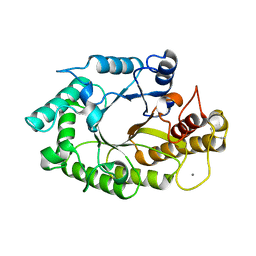 | | The 3-dimensional structure of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Taylor, E.J, Vincent, F, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
1W32
 
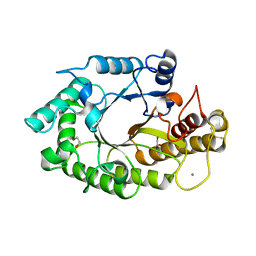 | | The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Andrews, S, Taylor, E.J, Pell, G.N, Vincent, F, Ducros, V.M.A, Davies, G.J, Lakey, J.H, Gilbert, H.J. | | Deposit date: | 2004-07-12 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
1W8U
 
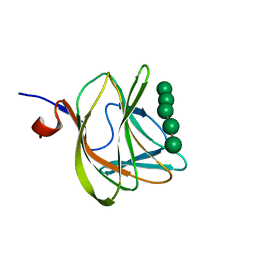 | | CBM29-2 mutant D83A complexed with mannohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
1W3L
 
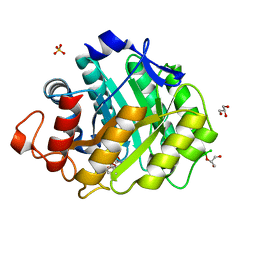 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOTRI DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
7OFX
 
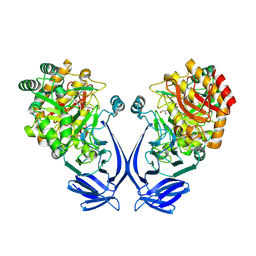 | | Crystal structure of a GH31 family sulfoquinovosidase mutant D455N from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | Alpha-glucosidase yihQ, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1WB5
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with syringate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6Z39
 
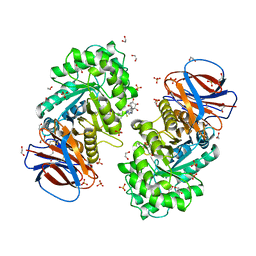 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY functionalised epoxide activity based probe | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[4-[4-[(12~{R})-2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-1,3-diaza-2$l^{4}-boratricyclo[7.3.0.0^{3,7}]dodeca-4,6,9-trien-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]-7-oxabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-19 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
2IW1
 
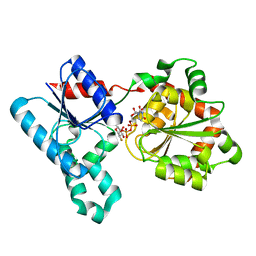 | | Crystal Structure of WaaG, a glycosyltransferase involved in lipopolysaccharide biosynthesis | | Descriptor: | LIPOPOLYSACCHARIDE CORE BIOSYNTHESIS PROTEIN RFAG, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Martinez-Fleites, C, Proctor, M, Roberts, S, Bolam, D.N, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2006-06-23 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights Into the Synthesis of Lipopolysaccharide and Antibiotics Through the Structures of Two Retaining Glycosyltransferases from Family Gt4
Chem.Biol., 13, 2006
|
|
6Z3I
 
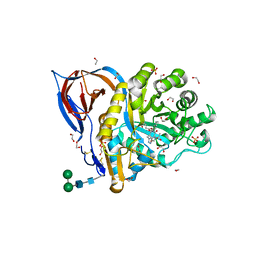 | | Structure of recombinant beta-glucocerebrosidase in complex with bifunctional cyclophellitol aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2020-05-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis and Structural Analysis of Glucocerebrosidase Imaging Agents.
Chemistry, 27, 2021
|
|
7OFY
 
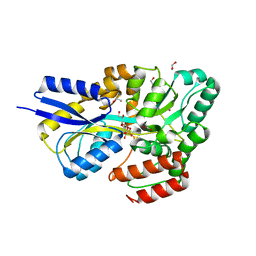 | | Crystal structure of SQ binding protein from Agrobacterium tumefaciens in complex with sulfoquinovosyl glycerol (SQGro) | | Descriptor: | 1,2-ETHANEDIOL, Sulfoquinovosyl binding protein, [(2S,3S,4S,5R,6S)-6-[(2R)-2,3-bis(oxidanyl)propoxy]-3,4,5-tris(oxidanyl)oxan-2-yl]methanesulfonic acid | | Authors: | Jarva, M.A, Sharma, M, Goddard-Borger, E.D, Davies, G.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OH2
 
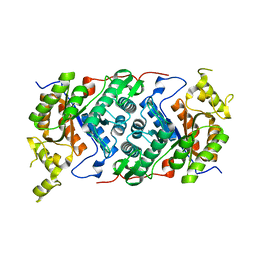 | |
2J7G
 
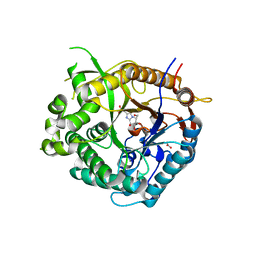 | | Beta-glucosidase from Thermotoga maritima in complex with methyl acetic acid-substituted glucoimidazole | | Descriptor: | ACETATE ION, BETA-GLUCOSIDASE A, CALCIUM ION, ... | | Authors: | Gloster, T.M, Zechel, D, Vasella, A, Davies, G.J. | | Deposit date: | 2006-10-07 | | Release date: | 2006-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Glycosidase Inhibition: An Assessment of the Binding of 18 Putative Transition-State Mimics.
J.Am.Chem.Soc., 129, 2007
|
|
1W8T
 
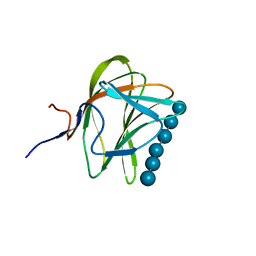 | | CBM29-2 mutant K74A complexed with cellulohexaose: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | NON CATALYTIC PROTEIN 1, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-09-28 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
6Z5Y
 
 | |
7ODJ
 
 | | Exo-mannosidase from Cellvibrio mixtus bound to N-alkyl mannocyclophellitol aziridine | | Descriptor: | (1R,2S,3R,4S,5R,6R)-5-(8-azidooctylamino)-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Man5A, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
1W18
 
 | | Crystal Structure of levansucrase from Gluconacetobacter diazotrophicus | | Descriptor: | LEVANSUCRASE, SULFATE ION | | Authors: | Martinez-Fleites, C, Ortiz-Lombardia, M, Pons, T, Tarbouriech, N, Taylor, E.J, Hernandez, L, Davies, G.J. | | Deposit date: | 2004-06-16 | | Release date: | 2005-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Levansucrase from the Gram- Negative Bacterium Gluconacetobacter Diazotrophicus.
Biochem.J., 390, 2005
|
|
1W2V
 
 | | The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Andrews, S, Taylor, E.J, Pell, G.N, Vincent, F, Ducros, V.M.A, Davies, G.J, Lakey, J.H, Glbert, H.J. | | Deposit date: | 2004-07-09 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
1W3H
 
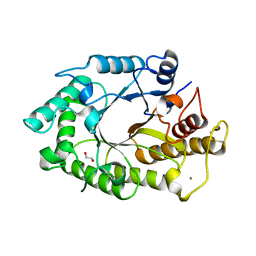 | | The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, ENDO-1,4-BETA-XYLANASE A PRECURSOR | | Authors: | Andrews, S, Taylor, E.J, Pell, G.N, Vincent, F, Ducros, V.M.A, Davies, G.J, Lakey, J.H, Glbert, H.J. | | Deposit date: | 2004-07-15 | | Release date: | 2004-09-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Use of Forced Protein Evolution to Investigate and Improve Stability of Family 10 Xylanases: The Production of Ca2+-Independent Stable Xylanases
J.Biol.Chem., 279, 2004
|
|
7NWV
 
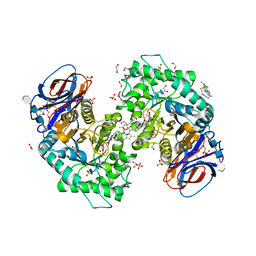 | | Structure of recombinant human beta-glucocerebrosidase in complex with BODIPY Tagged Cyclophellitol activity based probe | | Descriptor: | (1~{S},2~{R},3~{R},4~{S},5~{S})-4-[[4-[4-[2,2-bis(fluoranyl)-4,6,10,12-tetramethyl-3-aza-1-azonia-2-boranuidatricyclo[7.3.0.0^{3,7}]dodeca-1(12),4,6,8,10-pentaen-8-yl]butyl]-1,2,3-triazol-1-yl]methyl]cyclohexane-1,2,3,5-tetrol, 1,2-ETHANEDIOL, 1-deoxy-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Fluorescence polarisation activity-based protein profiling for the identification of deoxynojirimycin-type inhibitors selective for lysosomal retaining alpha- and beta-glucosidases.
Chem Sci, 14, 2023
|
|
6ZPV
 
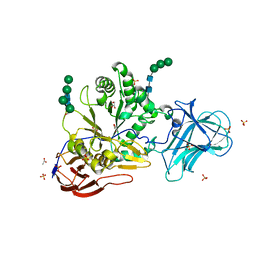 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZQ0
 
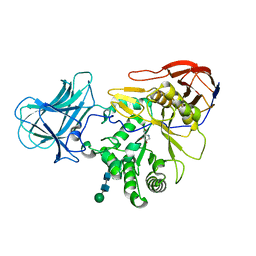 | | Structure of a-l-AraAZI-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPW
 
 | | Structure of Unliganded MgGH51 a-L-Arabinofuranosidase Crystal Type 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.329 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6ZPZ
 
 | | Structure of a-l-AraCS-Bound MgGH51 a-L-Arabinofuranosidase Crystal Type 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, MgGH51, ... | | Authors: | McGregor, N.G.S, Davies, G.J. | | Deposit date: | 2020-07-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure of a GH51 alpha-L-arabinofuranosidase from Meripilus giganteus: conserved substrate recognition from bacteria to fungi.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
1WB6
 
 | | S954A mutant of the feruloyl esterase module from clostridium thermocellum complexed with vanillate | | Descriptor: | ACETATE ION, CADMIUM ION, ENDO-1,4-BETA-XYLANASE Y, ... | | Authors: | Tarbouriech, N, Prates, J.A, Fontes, C, Davies, G.J. | | Deposit date: | 2004-10-30 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Determinants of Substrate Specificity in the Feruloyl Esterase Module of Xylanase 10B from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1W90
 
 | | CBM29-2 mutant D114A: Probing the Mechanism of Ligand Recognition by Family 29 Carbohydrate Binding Modules | | Descriptor: | 1,2-ETHANEDIOL, NON-CATALYTIC PROTEIN 1, SODIUM ION | | Authors: | Flint, J, Bolam, D.N, Nurizzo, D, Taylor, E.J, Williamson, M.P, Walters, C, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2004-10-01 | | Release date: | 2005-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the Mechanism of Ligand Recognition in Family 29 Carbohydrate-Binding Modules
J.Biol.Chem., 280, 2005
|
|
