3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | 分子名称: | GLYCEROL, YetF protein | | 著者 | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-02-04 | | 公開日 | 2008-02-19 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
4ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/OLOMOUCINE | | 分子名称: | EXTRACELLULAR REGULATED KINASE 2, OLOMOUCINE, SULFATE ION | | 著者 | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | 登録日 | 1998-07-09 | | 公開日 | 1999-07-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | 分子名称: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | 著者 | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | 登録日 | 2012-06-14 | | 公開日 | 2012-06-27 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
3SK0
 
 | | structure of Rhodococcus rhodochrous haloalkane dehalogenase DhaA mutant DhaA12 | | 分子名称: | CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Lahoda, M, Stsiapanava, A, Mesters, J, Koudelakova, T, Damborsky, J, Kuta-Smatanova, I. | | 登録日 | 2011-06-22 | | 公開日 | 2012-06-27 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Dynamics and hydration explain failed functional transformation in dehalogenase design.
Nat.Chem.Biol., 10, 2014
|
|
2OZT
 
 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | 分子名称: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | 著者 | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-02-27 | | 公開日 | 2007-03-13 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BT0
 
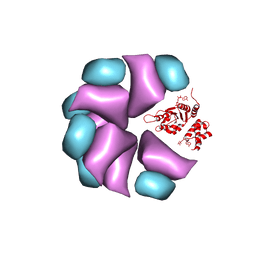 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | 著者 | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | 登録日 | 2013-06-12 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (17 Å) | | 主引用文献 | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BT1
 
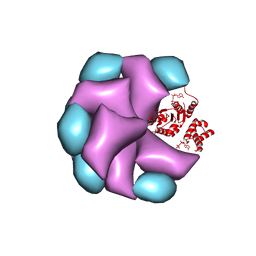 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | 著者 | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | 登録日 | 2013-06-12 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (16 Å) | | 主引用文献 | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BS1
 
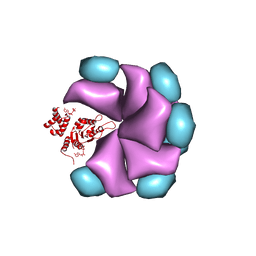 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR (NTRC FAMILY) | | 著者 | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | 登録日 | 2013-06-06 | | 公開日 | 2013-07-03 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (18 Å) | | 主引用文献 | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3FM3
 
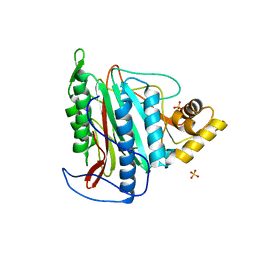 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 | | 分子名称: | FE (III) ION, Methionine aminopeptidase 2, SULFATE ION | | 著者 | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-12-19 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
3FMQ
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor fumagillin bound | | 分子名称: | FE (III) ION, FUMAGILLIN, Methionine aminopeptidase 2, ... | | 著者 | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2008-12-22 | | 公開日 | 2009-01-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
2P3Z
 
 | | Crystal structure of L-Rhamnonate dehydratase from Salmonella typhimurium | | 分子名称: | L-rhamnonate dehydratase, SODIUM ION | | 著者 | Malashkevich, V.N, Sauder, J.M, Dickey, M, Adams, J.M, Burley, S.K, Wasserman, S.R, Gerlt, J, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-03-10 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of L-Rhamnonate Dehydratase from Salmonella Typhimurium Lt2
To be Published
|
|
3RK4
 
 | | Structure of Rhodococcus rhodochrous haloalkane dehalogenase mutant DhaA31 | | 分子名称: | CHLORIDE ION, Haloalkane dehalogenase | | 著者 | Lahoda, M, Stsiapanava, A, Mesters, J, Chaloupkova, R, Damborsky, J, Kuta Smatanova, I. | | 登録日 | 2011-04-17 | | 公開日 | 2012-04-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | Crystallographic analysis of 1,2,3-trichloropropane biodegradation by the haloalkane dehalogenase DhaA31.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
