2F1F
 
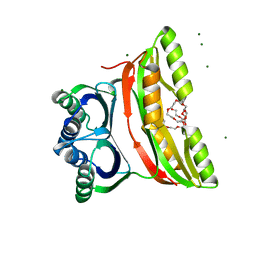 | | Crystal structure of the regulatory subunit of acetohydroxyacid synthase isozyme III from E. coli | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, Acetolactate synthase isozyme III small subunit, MAGNESIUM ION, ... | | Authors: | Kaplun, A, Vyazmensky, M, Barak, Z, Chipman, D.M, Shaanan, B. | | Deposit date: | 2005-11-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Regulatory Subunit of Acetohydroxyacid Synthase Isozyme III from Escherichia coli.
J.Mol.Biol., 357, 2006
|
|
2JYF
 
 | |
1XU8
 
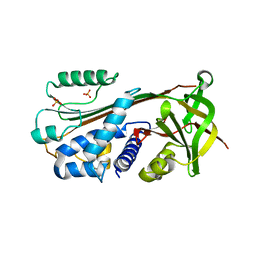 | | The 2.8 A structure of a tumour suppressing serpin | | Descriptor: | Maspin, SULFATE ION | | Authors: | Irving, J.A, Law, R.H, Ruzyla, K, Bashtannyk-Puhalovich, T.A, Kim, N, Worrall, D.M, Rossjohn, J, Whisstock, J.C. | | Deposit date: | 2004-10-25 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The high resolution crystal structure of the human tumor suppressor maspin reveals a novel conformational switch in the G-helix.
J.Biol.Chem., 280, 2005
|
|
2G5R
 
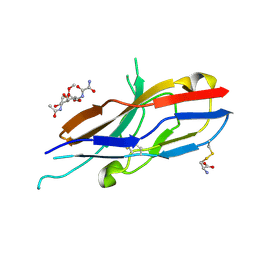 | | Crystal structure of Siglec-7 in complex with methyl-9-(aminooxalyl-amino)-9-deoxyNeu5Ac (oxamido-Neu5Ac) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYSTEINE, Sialic acid-binding Ig-like lectin 7, ... | | Authors: | Attrill, H, Crocker, P.R, van Aalten, D.M. | | Deposit date: | 2006-02-23 | | Release date: | 2006-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of siglec-7 in complex with sialosides: leads for rational structure-based inhibitor design.
Biochem.J., 397, 2006
|
|
1E84
 
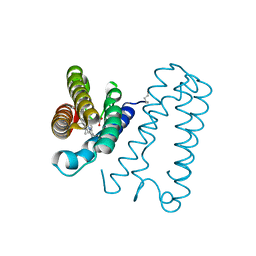 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
1E86
 
 | | Cytochrome c' from Alcaligenes xylosoxidans - reduced structure with CO bound to distal side of heme | | Descriptor: | CARBON MONOXIDE, CYTOCHROME C', HEME C | | Authors: | Lawson, D.M, Stevenson, C.E.M, Andrew, C.R, Eady, R.R. | | Deposit date: | 2000-09-15 | | Release date: | 2000-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Unprecedented Proximal Binding of Nitric Oxide to Heme: Implications for Guanylate Cyclase
Embo J., 19, 2000
|
|
2GBJ
 
 | |
2GBN
 
 | |
1Z16
 
 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound leucine | | Descriptor: | CADMIUM ION, LEUCINE, Leu/Ile/Val-binding protein | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
2GBM
 
 | | Crystal Structure of the 35-36 8 Glycine Insertion Mutant of Ubiquitin | | Descriptor: | ARSENIC, Ubiquitin | | Authors: | Ferraro, D.M, Ferraro, D.J, Ramaswamy, S, Robertson, A.D. | | Deposit date: | 2006-03-10 | | Release date: | 2006-05-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structures of Ubiquitin Insertion Mutants Support Site-specific Reflex Response to Insertions Hypothesis.
J.Mol.Biol., 359, 2006
|
|
2GBK
 
 | |
1FBL
 
 | | STRUCTURE OF FULL-LENGTH PORCINE SYNOVIAL COLLAGENASE (MMP1) REVEALS A C-TERMINAL DOMAIN CONTAINING A CALCIUM-LINKED, FOUR-BLADED BETA-PROPELLER | | Descriptor: | CALCIUM ION, FIBROBLAST (INTERSTITIAL) COLLAGENASE (MMP-1), N-[3-(N'-HYDROXYCARBOXAMIDO)-2-(2-METHYLPROPYL)-PROPANOYL]-O-TYROSINE-N-METHYLAMIDE, ... | | Authors: | Li, J, Brick, P, Blow, D.M. | | Deposit date: | 1995-04-24 | | Release date: | 1996-01-29 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller.
Structure, 3, 1995
|
|
1FD3
 
 | | HUMAN BETA-DEFENSIN 2 | | Descriptor: | BETA-DEFENSIN 2, SULFATE ION | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2000-07-19 | | Release date: | 2000-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The structure of human beta-defensin-2 shows evidence of higher order oligomerization.
J.Biol.Chem., 275, 2000
|
|
2HEV
 
 | | Crystal structure of the complex between OX40L and OX40 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 4, Tumor necrosis factor receptor superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
2HEW
 
 | | The X-ray crystal structure of murine OX40L | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tumor necrosis factor ligand superfamily member 4 | | Authors: | Hymowitz, S.G, Compaan, D.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
1XYR
 
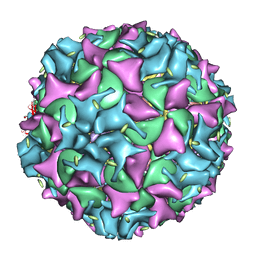 | | Poliovirus 135S cell entry intermediate | | Descriptor: | Genome polyprotein, Coat protein VP1, Coat protein VP2, ... | | Authors: | Bubeck, D, Filman, D.J, Cheng, N, Steven, A.C, Hogle, J.M, Belnap, D.M. | | Deposit date: | 2004-11-10 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes.
J.Virol., 79, 2005
|
|
1FYY
 
 | | HPRT GENE MUTATION HOTSPOT WITH A BPDE2(10R) ADDUCT | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 5'-D(*TP*GP*CP*CP*CP*TP*TP*GP*AP*CP*TP*A)-3', HPRT DNA WITH BENZO[A]PYRENE-ADDUCTED DA7 | | Authors: | Volk, D.E, Rice, J.S, Luxon, B.A, Yeh, H.J.C, Liang, C, Xie, G, Sayer, J.M, Jerina, D.M, Gorenstein, D.G. | | Deposit date: | 2000-10-03 | | Release date: | 2000-12-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR evidence for syn-anti interconversion of a trans opened (10R)-dA adduct of benzo[a]pyrene (7S,8R)-diol (9R,10S)-epoxide in a DNA duplex.
Biochemistry, 39, 2000
|
|
1Z18
 
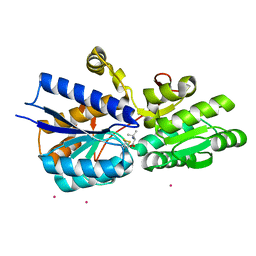 | | Crystal structure analysis of periplasmic Leu/Ile/Val-binding protein with bound valine | | Descriptor: | CADMIUM ION, Leu/Ile/Val-binding protein, VALINE | | Authors: | Trakhanov, S.D, Vyas, N.K, Kristensen, D.M, Ma, J, Quiocho, F.A. | | Deposit date: | 2005-03-03 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-free and -bound structures of the binding protein (LivJ) of the Escherichia coli ABC leucine/isoleucine/valine transport system: trajectory and dynamics of the interdomain rotation and ligand specificity.
Biochemistry, 44, 2005
|
|
2K1N
 
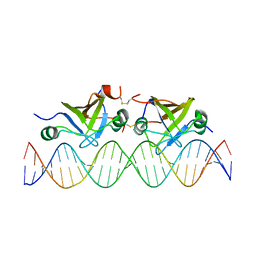 | | DNA bound structure of the N-terminal domain of AbrB | | Descriptor: | AbrB family transcriptional regulator, DNA (25-MER) | | Authors: | Cavanagh, J, Bobay, B.G, Sullivan, D.M, Thompson, R.J. | | Deposit date: | 2008-03-10 | | Release date: | 2008-11-11 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Insights into the Nature of DNA Binding of AbrB-like Transcription Factors
Structure, 16, 2008
|
|
1ZI0
 
 | |
1YUX
 
 | | Mixed valant state of nigerythrin | | Descriptor: | FE (II) ION, FE (III) ION, Nigerythrin | | Authors: | Iyer, R.B, Silaghi-Dumitrescu, R, Kurtz, D.M, Lanzilotta, W.N. | | Deposit date: | 2005-02-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins.
J.Biol.Inorg.Chem., 10, 2005
|
|
2JX5
 
 | | Solution structure of the ubiquitin domain N-terminal to the S27a ribosomal subunit of Giardia lamblia | | Descriptor: | GlUb(S27a) | | Authors: | Catic, A, Sun, Z.J, Ratner, D.M, Misaghi, S, Spooner, E, Samuelson, J, Wagner, G, Ploegh, H.L. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sequence and structure evolved separately in a ribosomal ubiquitin variant
EMBO J., 26, 2007
|
|
1CZH
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-03 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1CZR
 
 | | COMPARISONS OF WILD TYPE AND MUTANT FLAVODOXINS FROM ANACYSTIS NIDULANS. STRUCTURAL DETERMINANTS OF THE REDOX POTENTIALS. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Hoover, D.M, Drennan, C.L, Metzger, A.L, Osborne, C, Weber, C.H, Pattridge, K.A, Ludwig, M.L. | | Deposit date: | 1999-09-07 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparisons of wild-type and mutant flavodoxins from Anacystis nidulans. Structural determinants of the redox potentials.
J.Mol.Biol., 294, 1999
|
|
1XZ0
 
 | | Crystal structure of CD1a in complex with a synthetic mycobactin lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(HYDROXY-HEXADECANOYL-AMINO)-2-{[(4S)-2-(2-HYDROXY-PHENYL)-4,5-DIHYDRO-OXAZOLE-4-CARBONYL]-AMINO}-HEXANOIC ACID 2-[(3S)-1-(TERT-BUTYL-DIPHENYL-SILANYLOXY)-2-OXO-AZEPAN-3-YLCARBAMOYL]-(1S)-1-METHYL-ETHYL ESTER, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Crispin, M.D, Bowden, T.A, Young, D.C, Cheng, T.Y, Hu, J, Costello, C.E, Miller, M.J, Moody, D.B, Wilson, I.A. | | Deposit date: | 2004-11-11 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of Lipopeptide Presentation by CD1a.
Immunity, 22, 2005
|
|
