6IBU
 
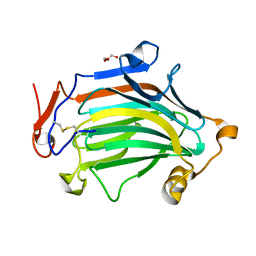 | | Apo Crh5 transglycosylase | | Descriptor: | GLYCEROL, Probable glycosidase crf1 | | Authors: | Bartual, S.G, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
1H9T
 
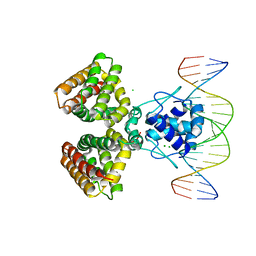 | | FADR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI IN COMPLEX WITH FADB OPERATOR | | Descriptor: | 5'-D(*CP*AP*TP*CP*TP*GP*GP*TP*AP*CP*GP*AP* CP*CP*AP*GP*AP*TP*C)-3', 5'-D(*GP*AP*TP*CP*TP*GP*GP*TP*CP*GP*TP*AP* CP*CP*AP*GP*AP*TP*G)-3', CHLORIDE ION, ... | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-19 | | Release date: | 2001-04-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
1GSV
 
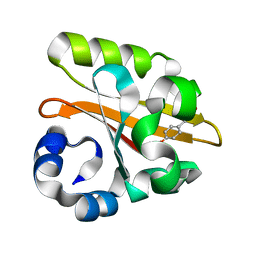 | | Crystal structure of the P65 crystal form of photoactive yellow protein G47S mutant | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1HJV
 
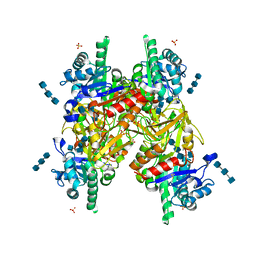 | | Crystal structure of hcgp-39 in complex with chitin tetramer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes.
J.Biol.Chem., 278, 2003
|
|
1HJX
 
 | | Ligand-induced signalling and conformational change of the 39 kD glycoprotein from human articular chondrocytes | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE-3 LIKE PROTEIN 1, GLYCEROL, ... | | Authors: | Houston, D.R, Recklies, A.D, Krupa, J.C, Van Aalten, D.M.F. | | Deposit date: | 2003-02-28 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Ligand-Induced Conformational Change of the 39-kDa Glycoprotein from Human Articular Chondrocytes
J.Biol.Chem., 278, 2003
|
|
1HNU
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1, PERRHENATE | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
6JIK
 
 | | Aspergillus fumigatus Rho1 GsGTP | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Rho GTPase Rho1 | | Authors: | Bartual, S.G, Van Aalten, D.M.F. | | Deposit date: | 2019-02-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Aspergillus fumigatus Rho1 gSGTP complex
To Be Published
|
|
1HKI
 
 | | Crystal structure of human chitinase in complex with glucoallosamidin B | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOTRIOSIDASE, METHYL N-ACETYL ALLOSAMINE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
1HKJ
 
 | | Crystal structure of human chitinase in complex with methylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-6-O-methyl-alpha-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITOTRIOSIDASE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
1HKK
 
 | | High resoultion crystal structure of human chitinase in complex with allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITOTRIOSIDASE-1, ... | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase.
J.Biol.Chem., 278, 2003
|
|
1HNO
 
 | | CRYSTAL STRUCTURE OF PEROXISOMAL DELTA3-DELTA2-ENOYL-COA ISOMERASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 1,2-ETHANEDIOL, D3,D2-ENOYL COA ISOMERASE ECI1 | | Authors: | Mursula, A.M, van Aalten, D.M.F, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 2000-12-08 | | Release date: | 2001-06-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of delta(3)-delta(2)-enoyl-CoA isomerase.
J.Mol.Biol., 309, 2001
|
|
1HKM
 
 | | High resolution crystal structure of human chitinase in complex with demethylallosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, CHITOTRIOSIDASE, METHYL N-ACETYL ALLOSAMINE | | Authors: | Rao, F.V, Houston, D.R, Boot, R.G, Aerts, J.M.F.G, Sakuda, S, Van Aalten, D.M.F. | | Deposit date: | 2003-03-10 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structures of Allosamidin Derivatives in Complex with Human Macrophage Chitinase
J.Biol.Chem., 278, 2003
|
|
6HBT
 
 | |
7EAG
 
 | | Crystal structure of the RAGATH-18 k-turn | | Descriptor: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2021-03-07 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
 | |
6HBX
 
 | |
6IBW
 
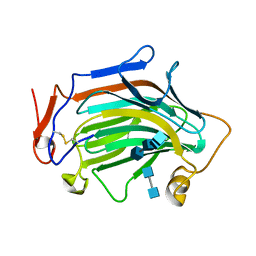 | | Crh5 transglycosylase in complex with NAG | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Probable glycosidase crf1 | | Authors: | Fang, W, Bartual, S.G, van Aalten, D.M.F. | | Deposit date: | 2018-12-01 | | Release date: | 2019-02-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanisms of redundancy and specificity of the Aspergillus fumigatus Crh transglycosylases.
Nat Commun, 10, 2019
|
|
6R47
 
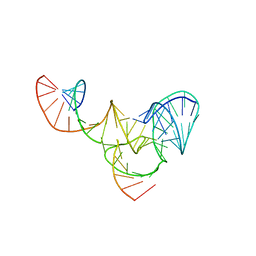 | |
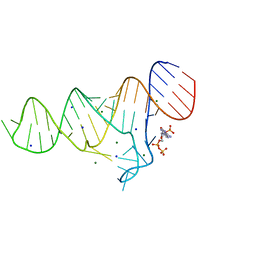
6TFG
 
 | |
6TF1
 
 | |
6TFF
 
 | |
6TF0
 
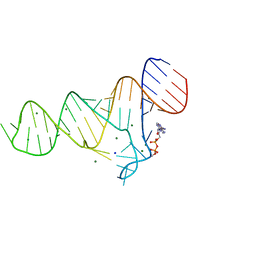 | | Crystal structure of the ADP-binding domain of the NAD+ riboswitch with Nicotinamide adenine dinucleotide, reduced (NADH) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Chains: A, MAGNESIUM ION, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and ligand binding of the ADP-binding domain of the NAD+ riboswitch.
Rna, 26, 2020
|
|
6TF3
 
 | |
6TFE
 
 | |
6HC5
 
 | |
