4LKA
 
 | | Crystal Structure of MOZ double PHD finger histone H3K9ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LLB
 
 | | Crystal Structure of MOZ double PHD finger histone H3K14ac complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
5CQ1
 
 | | Disproportionating enzyme 1 from Arabidopsis - cycloamylose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
3RZC
 
 | | Structure of the self-antigen iGb3 bound to mouse CD1d and in complex with the iNKT TCR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Yu, E.D, Girardi, E, Wang, J, Zajonc, D.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cutting Edge: Structural Basis for the Recognition of {beta}-Linked Glycolipid Antigens by Invariant NKT Cells.
J.Immunol., 187, 2011
|
|
5DCW
 
 | | Iridoid synthase from Catharanthus roseus - ligand free structure | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
5CSY
 
 | | Disproportionating enzyme 1 from Arabidopsis - acarbose soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
4LDB
 
 | |
3S7O
 
 | | Crystal Structure of the Infrared Fluorescent D207H variant of Deinococcus Bacteriophytochrome chromophore binding domain at 1.24 angstrom resolution | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome, GLYCEROL | | Authors: | Forest, K.T, Auldridge, M.E, Satyshur, K.A, Anstrom, D.M. | | Deposit date: | 2011-05-26 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein.
J.Biol.Chem., 287, 2012
|
|
9MHB
 
 | | Crystal Structure of Human P38 alpha MAPK In Complex with MW01-14-064SRM | | Descriptor: | (5P)-N,N-dimethyl-5-(naphthalen-1-yl)-6-(pyridin-4-yl)pyrazin-2-amine, 1,2-ETHANEDIOL, 4-[3-(4-FLUOROPHENYL)-1H-PYRAZOL-4-YL]PYRIDINE, ... | | Authors: | Minasov, G, Tokars, V.L, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-12-11 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Human P38 alpha MAPK In Complex with MW01-14-064SRM
To Be Published
|
|
3SDE
 
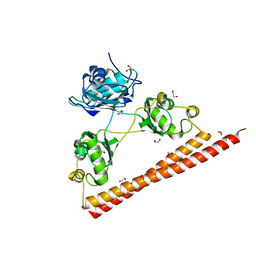 | | Crystal structure of a paraspeckle-protein heterodimer, PSPC1/NONO | | Descriptor: | 1,2-ETHANEDIOL, Non-POU domain-containing octamer-binding protein, Paraspeckle component 1 | | Authors: | Passon, D.M, Lee, M, Bond, C.S. | | Deposit date: | 2011-06-09 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
9MIU
 
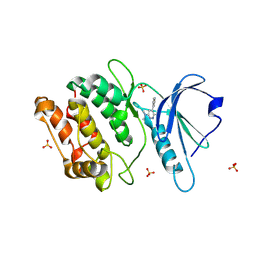 | | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound MW01-9-039SRM. | | Descriptor: | 6-chloro-N,N-dimethyl-5-(pyridin-4-yl)pyridazin-3-amine, Death-associated protein kinase 1, SULFATE ION | | Authors: | Minasov, G, Winsor, J, Roy, S.M, Watterson, D.M, Shuvalova, L. | | Deposit date: | 2024-12-13 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human DAPK1 Catalytic Subunit Complexed with Compound MW01-9-039SRM.
To Be Published
|
|
5CPT
 
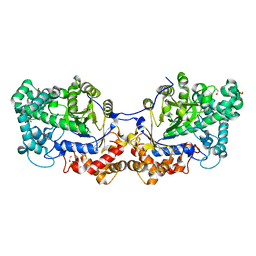 | | Disproportionating enzyme 1 from Arabidopsis - beta cyclodextrin soak | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase DPE1, chloroplastic/amyloplastic, ... | | Authors: | O'Neill, E.C, Stevenson, C.E.M, Tantanarat, K, Latousakis, D, Donaldson, M.I, Rejzek, M, Limpaseni, T, Smith, A.M, Field, R.A, Lawson, D.M. | | Deposit date: | 2015-07-21 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Dissection of the Maltodextrin Disproportionation Cycle of the Arabidopsis Plastidial Disproportionating Enzyme 1 (DPE1).
J.Biol.Chem., 290, 2015
|
|
4LD8
 
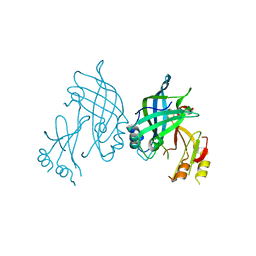 | |
4LJN
 
 | | Crystal Structure of MOZ double PHD finger | | Descriptor: | Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-05 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
4LK9
 
 | | Crystal Structure of MOZ double PHD finger histone H3 tail complex | | Descriptor: | Histone H3.1, Histone acetyltransferase KAT6A, ZINC ION | | Authors: | Dreveny, I, Deeves, S.E, Yue, B, Heery, D.M. | | Deposit date: | 2013-07-07 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The double PHD finger domain of MOZ/MYST3 induces alpha-helical structure of the histone H3 tail to facilitate acetylation and methylation sampling and modification.
Nucleic Acids Res., 42, 2014
|
|
9MUL
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with Compound 1, Glycine and Glutamate | | Descriptor: | 5-[(3-chlorobenzene-1-sulfonyl)methoxy]-6-methyl-N-[(pyridin-3-yl)methyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Shaffer, P.L, Duda, D.M. | | Deposit date: | 2025-01-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design, Synthesis, and Characterization of GluN2A Negative Allosteric Modulators Suitable for In Vivo Exploration.
J.Med.Chem., 68, 2025
|
|
9MUM
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with Compound 11, Glycine and Glutamate | | Descriptor: | 5-[2-(3-chlorophenyl)-2,2-difluoroethoxy]-N-[1-(pyrazin-2-yl)cyclopropyl]pyrazine-2-carboxamide, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Shaffer, P.L, Duda, D.M. | | Deposit date: | 2025-01-14 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design, Synthesis, and Characterization of GluN2A Negative Allosteric Modulators Suitable for In Vivo Exploration.
J.Med.Chem., 68, 2025
|
|
5CVS
 
 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltoheptaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Rashid, A.M, Syson, K, Koliwer-Brandl, H, van de Weerd, R, Stevenson, C.E.M, Batey, S.F.D, Miah, F, Alber, M, Ioerger, T.R, Chandra, G, Appelmelk, B.J, Nartowski, K.P, Khimyak, Y.Z, Lawson, D.M, Jacobs, W.R, Geurtsen, J, Kalscheuer, R, Bornemann, S. | | Deposit date: | 2015-07-27 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
9N1T
 
 | |
5DCU
 
 | | Iridoid synthase from Catharanthus roseus - ternary complex with NADP+ and triethylene glycol carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, Iridoid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Caputi, L, Kries, H, Stevenson, C.E.M, Kamileen, M.O, Sherden, N.H, Geu-Flores, F, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2015-08-24 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural determinants of reductive terpene cyclization in iridoid biosynthesis.
Nat.Chem.Biol., 12, 2016
|
|
3S6X
 
 | |
9M0D
 
 | | Cryo-EM structure of neurotensin receptor 1 in complex with beta-arrestin 1 | | Descriptor: | Beta-arrestin-1, Fab30 heavy chain, Fab30 light chain, ... | | Authors: | Sun, D.M, Li, X, Yuan, Q.N, Tian, C.L. | | Deposit date: | 2025-02-24 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Molecular mechanism of the arrestin-biased agonism of neurotensin receptor 1 by an intracellular allosteric modulator.
Cell Res., 35, 2025
|
|
3S6Z
 
 | |
9N2M
 
 | |
9N2O
 
 | |
