3ZFS
 
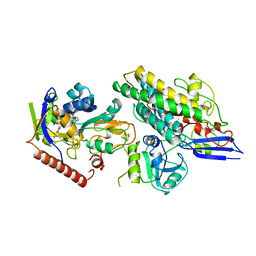 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
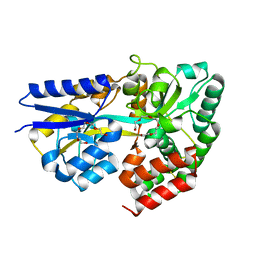
3ZP6
 
 | | INFLUENZA VIRUS (VN1194) H5 E190D mutant HA with LSTc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HAEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
3ZKL
 
 | | Structure of the xylo-oligosaccharide specific solute binding protein from Bifidobacterium animalis subsp. lactis Bl-04 in complex with xylotriose | | Descriptor: | PENTAETHYLENE GLYCOL, PUTATIVE SUGAR TRANSPORTER SOLUTE-BINDING PROTEIN, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Ejby, M, Vujicic-Zagar, A, Fredslund, F, Svensson, B, Slotboom, D.J, Abou Hachem, M. | | Deposit date: | 2013-01-23 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Basis for Arabinoxylo-Oligosaccharide Capture by the Probiotic Bifidobacterium Animalis Subsp. Lactis Bl-04
Mol.Microbiol., 90, 2013
|
|
3ZP0
 
 | | INFLUENZA VIRUS (VN1194) H5 HA with LSTa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HEMAGGLUTININ, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose | | Authors: | Liu, J, Stevens, D.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2013-02-26 | | Release date: | 2013-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Changes in the Hemagglutinin of H5N1 Viruses During Human Infection - Influence on Receptor Binding.
Virology, 447, 2013
|
|
8G66
 
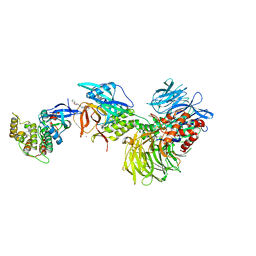 | | Structure with SJ3149 | | Descriptor: | (3S)-3-{5-[(1,2-benzoxazol-3-yl)amino]-1-oxo-1,3-dihydro-2H-isoindol-2-yl}piperidine-2,6-dione, Casein kinase I isoform alpha, DNA damage-binding protein 1, ... | | Authors: | Miller, D.J, Young, S.M, Fischer, M. | | Deposit date: | 2023-02-14 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structure of ternary complex with molecular glue targeting CK1A for degradation by the CRL4CRBN ubiquitin ligase
To Be Published
|
|
1ZOP
 
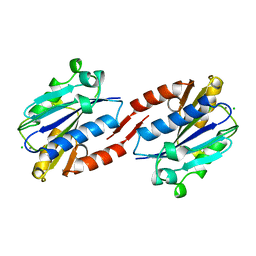 | | CD11A I-DOMAIN WITH BOUND MAGNESIUM ION | | Descriptor: | CHLORIDE ION, I-DOMAIN FRAGMENT OF LFA-1, MANGANESE (II) ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
1ZOO
 
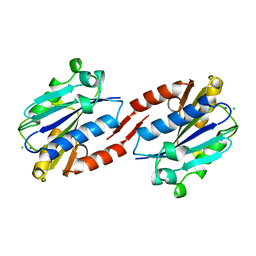 | | CD11A I-DOMAIN WITH BOUND MAGNESIUM ION | | Descriptor: | CHLORIDE ION, LEUKOCYTE ADHESION GLYCOPROTEIN, MAGNESIUM ION | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-21 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
1ZON
 
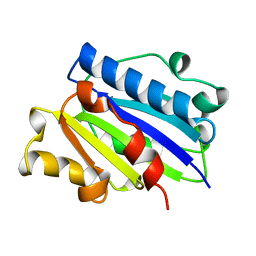 | | CD11A I-DOMAIN WITHOUT BOUND CATION | | Descriptor: | LEUKOCYTE ADHESION GLYCOPROTEIN | | Authors: | Leahy, D.J, Qu, A. | | Deposit date: | 1996-06-20 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The role of the divalent cation in the structure of the I domain from the CD11a/CD18 integrin.
Structure, 4, 1996
|
|
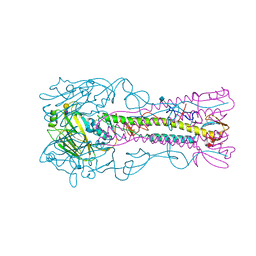
4A7F
 
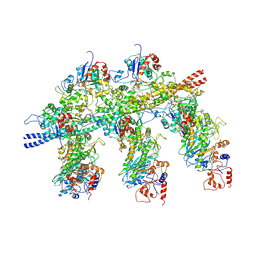 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 3) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
3UII
 
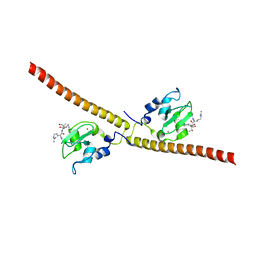 | |
3UP0
 
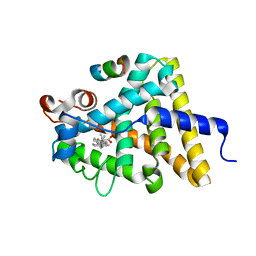 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-delta7-dafachronic acid | | Descriptor: | (5beta,14beta,17alpha,25S)-3-oxocholest-7-en-26-oic acid, Nuclear receptor coactivator 2, aceDAF-12 | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
4AE3
 
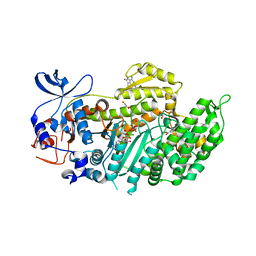 | | Crystal structure of ammosamide 272:myosin-2 motor domain complex | | Descriptor: | 1,2-ETHANEDIOL, ADP ORTHOVANADATE, AMMOSAMIDE 272, ... | | Authors: | Chinthalapudi, K, Heissler, S.M, Fenical, W, Manstein, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ammosamide Mediated Myosin Motor Activity Inhibition
To be Published
|
|
4A3O
 
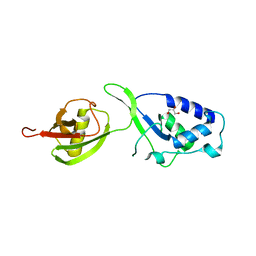 | | Crystal structure of the USP15 DUSP-UBL monomer | | Descriptor: | GLYCEROL, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | Elliott, P.R, Liu, H, Pastok, M.W, Grossmann, G.J, Rigden, D.J, Clague, M.J, Urbe, S, Barsukov, I.L. | | Deposit date: | 2011-10-03 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Variability of the Ubiquitin Specific Protease Dusp-Ubl Double Domains.
FEBS Lett., 585, 2011
|
|
433D
 
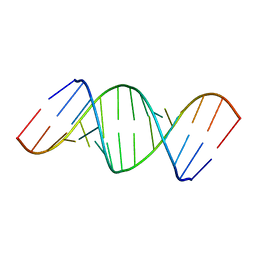 | |
484D
 
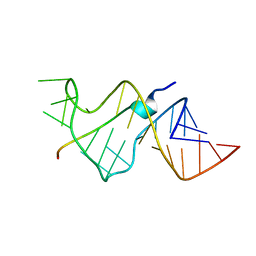 | | SOLUTION STRUCTURE OF HIV-1 REV PEPTIDE-RNA APTAMER COMPLEX | | Descriptor: | BASIC REV PEPTIDE, RNA APTAMER | | Authors: | Ye, X, Gorin, A.A, Frederick, R, Hu, W, Majumdar, A, Xu, W, Mclendon, G, Ellington, A, Patel, D.J. | | Deposit date: | 1999-08-02 | | Release date: | 1999-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | RNA architecture dictates the conformations of a bound peptide.
Chem.Biol., 6, 1999
|
|
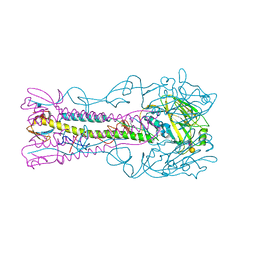
4A7L
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 1) | | Descriptor: | ACTIN, ALPHA SKELETON MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7N
 
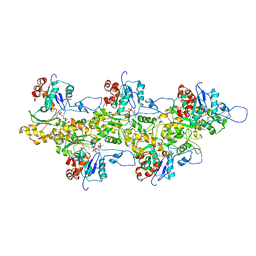 | | Structure of bare F-actin filaments obtained from the same sample as the Actin-Tropomyosin-Myosin Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, F-ACTIN | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
4A7H
 
 | | Structure of the Actin-Tropomyosin-Myosin Complex (rigor ATM 2) | | Descriptor: | ACTIN, ALPHA SKELETAL MUSCLE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Behrmann, E, Mueller, M, Penczek, P.A, Mannherz, H.G, Manstein, D.J, Raunser, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-08-01 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structure of the Rigor Actin-Tropomyosin-Myosin Complex.
Cell(Cambridge,Mass.), 150, 2012
|
|
7M3N
 
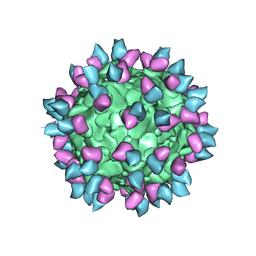 | |
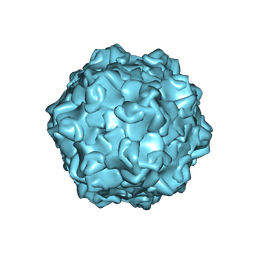
7M3L
 
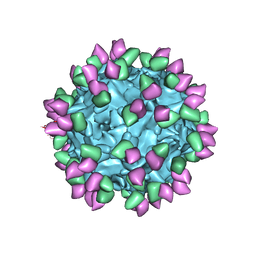 | |
7M3O
 
 | |
7M3M
 
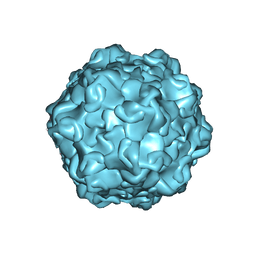 | |
3UP3
 
 | | Nuclear receptor DAF-12 from hookworm Ancylostoma ceylanicum in complex with (25S)-cholestenoic acid | | Descriptor: | (8alpha,10alpha,25S)-3-hydroxycholesta-3,5-dien-26-oic acid, 1,2-ETHANEDIOL, Nuclear receptor coactivator 2, ... | | Authors: | Zhi, X, Zhou, X.E, Melcher, K, Motola, D.L, Gelmedin, V, Hawdon, J, Kliewer, S.A, Mangelsdorf, D.J, Xu, H.E. | | Deposit date: | 2011-11-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structural Conservation of Ligand Binding Reveals a Bile Acid-like Signaling Pathway in Nematodes.
J.Biol.Chem., 287, 2012
|
|
3UVC
 
 | | MMP12 in a complex with the dimeric adduct: 5-(5-phenylhydantoin)-5-phenylhydantoin | | Descriptor: | (4R,4'S)-4,4'-diphenyl-4,4'-biimidazolidine-2,2',5,5'-tetrone, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Derbyshire, D.J, Danielson, H, Nystrum, S. | | Deposit date: | 2011-11-29 | | Release date: | 2013-01-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of fragments interacting with MMP-12
To be Published
|
|
4AK7
 
 | | Crystal structure of BpGH117_E303Q in complex with neoagarobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ANHYDRO-ALPHA-L-GALACTOSIDASE, ... | | Authors: | Hehemann, J.H, Smyth, L, Yadav, A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-02-21 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of Keystone Enzyme in Agar Hydrolysis Provides Insight Into the Degradation (of a Polysaccharide from) Red Seaweeds.
J.Biol.Chem., 287, 2012
|
|
