1OJV
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK1
 
 | | Decay accelerating factor (cd55) : the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK3
 
 | | Decay accelerating factor (cd55): the structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1OK9
 
 | | Decay accelerating factor (CD55): The structure of an intact human complement regulator. | | Descriptor: | ACETATE ION, CHLORIDE ION, COMPLEMENT DECAY-ACCELERATING FACTOR, ... | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, R.A, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-21 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1H2P
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H03
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1QOM
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH SWAPPED N-TERMINAL HOOK | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Crane, B.R, Rosenfeld, R.A, Arvai, A.S, Tainer, J.A, Stuehr, D.J, Getzoff, E.D. | | Deposit date: | 1999-11-15 | | Release date: | 1999-12-15 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | N-Terminal Domain Swapping and Metal Ion Binding in Nitric Oxide Synthase Dimerization
Embo J., 18, 1999
|
|
1H2Q
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I.G, Billington, J, Powell, R, Spiller, O.B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-08-13 | | Release date: | 2003-09-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1H04
 
 | | Human CD55 domains 3 & 4 | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, NICKEL (II) ION | | Authors: | Williams, P, Chaudhry, Y, Goodfellow, I, Billington, J, Spiller, B, Evans, D.J, Lea, S.M. | | Deposit date: | 2002-06-11 | | Release date: | 2003-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Cd55 Function. The Structure of Two Pathogen-Binding Domains at 1.7 A
J.Biol.Chem., 278, 2003
|
|
1IEN
 
 | | SOLUTION STRUCTURE OF TIA | | Descriptor: | PROTEIN TIA | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1DWW
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and dihydrobiopterin | | Descriptor: | 2-AMINO-6-(1,2-DIHYDROXY-PROPYL)-7,8-DIHYDRO-6H-PTERIDIN-4-ONE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1OJW
 
 | | Decay accelerating factor (CD55): the structure of an intact human complement regulator. | | Descriptor: | COMPLEMENT DECAY-ACCELERATING FACTOR, GLYCEROL, SULFATE ION | | Authors: | Lukacik, P, Roversi, P, White, J, Esser, D, Smith, G.P, Billington, J, Williams, P.A, Rudd, P.M, Wormald, M.R, Crispin, M.D.M, Radcliffe, C.M, Dwek, C.M, Evans, D.J, Morgan, B.P, Smith, R.A.G, Lea, S.M. | | Deposit date: | 2003-07-16 | | Release date: | 2004-01-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complement Regulation at the Molecular Level: The Structure of Decay-Accelerating Factor
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1DWX
 
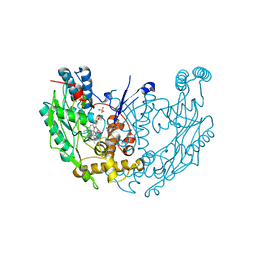 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and tetrahydrobiopterin | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DWV
 
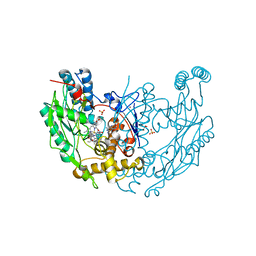 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER N-hydroxyarginine and 4-amino tetrahydrobiopterin | | Descriptor: | 2,4-DIAMINO-6-[2,3-DIHYDROXY-PROP-3-YL]-5,6,7,8-TETRAHYDROPTERIDINE, N-OMEGA-HYDROXY-L-ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1999-12-14 | | Release date: | 2000-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structures of the N(Omega)-Hydroxy-L-Arginine Complex of Inducible Nitric Oxide Synthase Oxygenase Dimer with Active Andinactive Pterins
Biochemistry, 39, 2000
|
|
1DNL
 
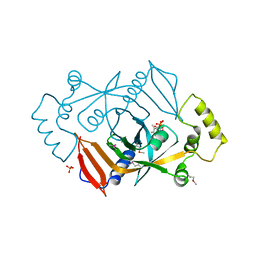 | | X-RAY STRUCTURE OF ESCHERICHIA COLI PYRIDOXINE 5'-PHOSPHATE OXIDASE COMPLEXED WITH FMN AT 1.8 ANGSTROM RESOLUTION | | Descriptor: | FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Safo, M.K, Mathews, I, Musayev, F.N, di Salvo, M.L, Thiel, D.J, Abraham, D.J, Schirch, V. | | Deposit date: | 1999-12-16 | | Release date: | 2000-01-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of Escherichia coli pyridoxine 5'-phosphate oxidase complexed with FMN at 1.8 A resolution.
Structure Fold.Des., 8, 2000
|
|
1IEO
 
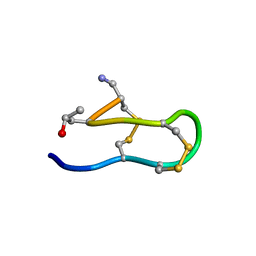 | | SOLUTION STRUCTURE OF MRIB-NH2 | | Descriptor: | PROTEIN MRIB-NH2 | | Authors: | Sharpe, I.A, Gehrmann, J, Loughnan, M.L, Thomas, L, Adams, D.A, Atkins, A, Palant, E, Craik, D.J, Adams, D.J, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2001-04-10 | | Release date: | 2002-04-03 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Two new classes of conopeptides inhibit the alpha1-adrenoceptor and noradrenaline transporter.
Nat.Neurosci., 4, 2001
|
|
1EWS
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE RABBIT KIDNEY DEFENSIN, RK-1 | | Descriptor: | RK-1 DEFENSIN | | Authors: | McManus, A.M, Dawson, N.F, Wade, J.D, Craik, D.J. | | Deposit date: | 2000-04-26 | | Release date: | 2001-05-02 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of RK-1: a novel alpha-defensin peptide.
Biochemistry, 39, 2000
|
|
2RD7
 
 | | Human Complement Membrane Attack Proteins Share a Common Fold with Bacterial Cytolysins | | Descriptor: | CHLORIDE ION, Complement component C8 alpha chain, Complement component C8 gamma chain | | Authors: | Slade, D.J, Lovelace, L.L, Chruszcz, M, Minor, W, Lebioda, L, Sodetz, J.M. | | Deposit date: | 2007-09-21 | | Release date: | 2008-05-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement.
J. Biol. Chem., 286, 2011
|
|
2MSQ
 
 | | Solution study of cBru9a | | Descriptor: | Conotoxin cBru9a | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2MSO
 
 | | Solution study of cGm9a | | Descriptor: | Conotoxin Gm9.1 | | Authors: | Akcan, M, Clark, R.J, Daly, N.L, Conibear, A.C, de Faoite, A.C, Heghinian, M.C, Adams, D.J, Mari, F, Craik, D.J. | | Deposit date: | 2014-08-05 | | Release date: | 2015-07-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Transforming conotoxins into cyclotides: Backbone cyclization of P-superfamily conotoxins.
Biopolymers, 104, 2015
|
|
2NS3
 
 | | Solution structure of ribbon BuIA | | Descriptor: | Alpha-conotoxin BuIA | | Authors: | Jin, A.H, Brandstaetter, H, Nevin, S.T, Tan, C.C, Clark, R.J, Adams, D.J, Alewood, P.F, Craik, D.J, Daly, N.L. | | Deposit date: | 2006-11-03 | | Release date: | 2007-09-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of alpha-conotoxin BuIA: influences of disulfide connectivity on structural dynamics
Bmc Struct.Biol., 7, 2007
|
|
6YCH
 
 | | Crystal structure of GcoA T296A bound to guaiacol | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Mallinson, S.J.B, Hinchen, D.J, Ellis, E.S, DuBois, J.L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCK
 
 | | Crystal structure of GcoA T296A bound to p-vanillin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCL
 
 | | Crystal structure of GcoA T296G bound to p-vanillin | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
6YCO
 
 | | Crystal structure of GcoA F169S bound to o-vanillin | | Descriptor: | 2-(hydroxymethyl)-6-methoxy-phenol, Aromatic O-demethylase, cytochrome P450 subunit, ... | | Authors: | Hinchen, D.J, Mallinson, S.J.B, Allen, M.D, Ellis, E.S, Beckham, G.T, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering a Cytochrome P450 for Demethylation of Lignin-Derived Aromatic Aldehydes.
Jacs Au, 1, 2021
|
|
