1SO0
 
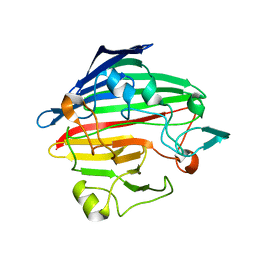 | | Crystal structure of human galactose mutarotase complexed with galactose | | Descriptor: | aldose 1-epimerase, beta-D-galactopyranose | | Authors: | Thoden, J.B, Timson, D.J, Reece, R.J, Holden, H.M. | | Deposit date: | 2004-03-12 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular structure of human galactose mutarotase
J.Biol.Chem., 279, 2004
|
|
2C9Z
 
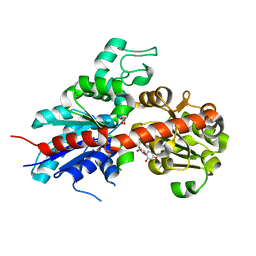 | | Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
4I4E
 
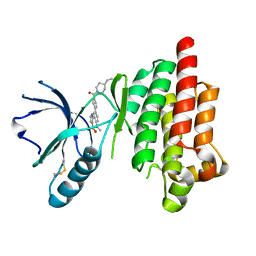 | | Structure of Focal Adhesion Kinase catalytic domain in complex with hinge binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, [4-(2-hydroxyethyl)piperidin-1-yl][4-(5-methyl-4,4-dioxido-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-yl)phenyl]methanone | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HZ3
 
 | | MthK pore crystallized in presence of TBSb | | Descriptor: | Calcium-gated potassium channel mthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The voltage-dependent gate in MthK potassium channels is located at the selectivity filter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2CHN
 
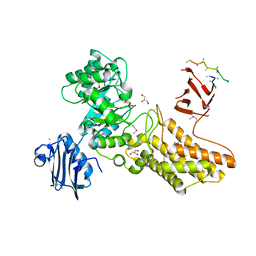 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity- NAG-thiazoline complex | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
1SBC
 
 | |
1T4F
 
 | | Structure of human MDM2 in complex with an optimized p53 peptide | | Descriptor: | SULFATE ION, Ubiquitin-protein ligase E3 Mdm2, optimized p53 peptide | | Authors: | Grasberger, B.L, Schubert, C, Koblish, H.K, Carver, T.E, Franks, C.F, Zhao, S.Y, Lu, T, LaFrance, L.V, Parks, D.J. | | Deposit date: | 2004-04-29 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells
J.Med.Chem., 48, 2005
|
|
1SU1
 
 | | Structural and biochemical characterization of Yfce, a phosphoesterase from E. coli | | Descriptor: | Hypothetical protein yfcE, SULFATE ION, ZINC ION | | Authors: | Miller, D.J, Shuvalova, L, Evdokimova, E, Savchenko, A, Yakunin, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-03-25 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and biochemical characterization of a novel Mn2+-dependent phosphodiesterase encoded by the yfcE gene.
Protein Sci., 16, 2007
|
|
4HYO
 
 | | Crystal Structure of MthK Pore | | Descriptor: | Calcium-gated potassium channel mthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2012-11-13 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The voltage-dependent gate in MthK potassium channels is located at the selectivity filter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
1T6O
 
 | | Nucleocapsid-binding domain of the measles virus P protein (amino acids 457-507) in complex with amino acids 486-505 of the measles virus N protein | | Descriptor: | linker, phosphoprotein | | Authors: | Kingston, R.L, Hamel, D.J, Gay, L.S, Dahlquist, F.W, Matthews, B.W. | | Deposit date: | 2004-05-06 | | Release date: | 2004-08-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the attachment of a paramyxoviral polymerase to its template.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
179D
 
 | |
186D
 
 | |
4W6A
 
 | |
1F40
 
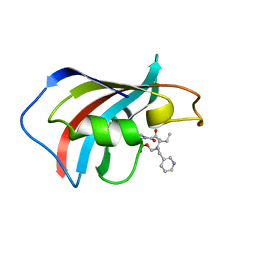 | | SOLUTION STRUCTURE OF FKBP12 COMPLEXED WITH GPI-1046, A NEUROTROPHIC LIGAND | | Descriptor: | (2S)-[3-PYRIDYL-1-PROPYL]-1-[3,3-DIMETHYL-1,2-DIOXOPENTYL]-2-PYRROLIDINECARBOXYLATE, FK506 BINDING PROTEIN (FKBP12) | | Authors: | Sich, C, Improta, S, Cowley, D.J, Guenet, C, Merly, J.P, Teufel, M, Saudek, V. | | Deposit date: | 2000-06-07 | | Release date: | 2000-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a neurotrophic ligand bound to FKBP12 and its effects on protein dynamics.
Eur.J.Biochem., 267, 2000
|
|
1A4T
 
 | | SOLUTION STRUCTURE OF PHAGE P22 N PEPTIDE-BOX B RNA COMPLEX, NMR, 20 STRUCTURES | | Descriptor: | 20-MER BASIC PEPTIDE, BOXB RNA | | Authors: | Cai, Z, Gorin, A.A, Frederick, R, Ye, X, Hu, W, Majumdar, A, Kettani, A, Patel, D.J. | | Deposit date: | 1998-02-04 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of P22 transcriptional antitermination N peptide-boxB RNA complex.
Nat.Struct.Biol., 5, 1998
|
|
1A6H
 
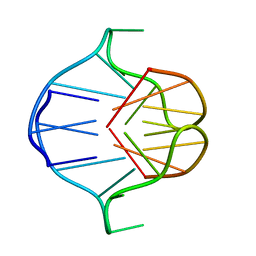 | |
4W6H
 
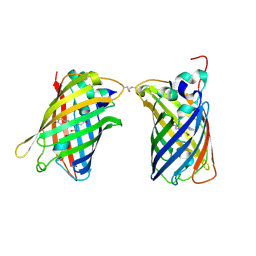 | |
7OEU
 
 | | Model of open pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum encapsulated ferritin, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
1AM0
 
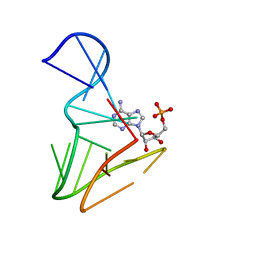 | | AMP RNA APTAMER COMPLEX, NMR, 8 STRUCTURES | | Descriptor: | ADENOSINE MONOPHOSPHATE, RNA APTAMER | | Authors: | Jiang, F, Kumar, R.A, Jones, R.A, Patel, D.J. | | Deposit date: | 1997-06-19 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of RNA Folding and Recognition in an AMP-RNA Aptamer Complex
Nature, 382, 1996
|
|
1F4U
 
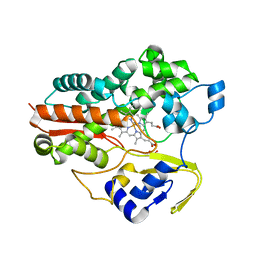 | | THERMOPHILIC P450: CYP119 FROM SULFOLOBUS SOLFACTARICUS | | Descriptor: | CYTOCHROME P450 119, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yano, J.K, Koo, L.S, Schuller, D.J, Li, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2000-06-09 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus.
J.Biol.Chem., 275, 2000
|
|
4W6L
 
 | |
4W6P
 
 | |
2KUX
 
 | |
4W73
 
 | |
199D
 
 | | Solution structure of the monoalkylated mitomycin c-DNA complex | | Descriptor: | CARBAMIC ACID 2,6-DIAMINO-5-METHYL-4,7-DIOXO-2,3,4,7-TETRAHYDRO-1H-3A-AZA-CYCLOPENTA[A]INDEN-8-YLMETHYL ESTER, DNA (5'-D(*(DI)P*CP*AP*CP*GP*TP*CP*(DI)P*T)-3'), DNA (5'-D(*AP*CP*GP*AP*CP*GP*TP*GP*C)-3') | | Authors: | Sastry, M, Fiala, R, Lipman, R, Tomasz, M, Patel, D.J. | | Deposit date: | 1994-12-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-03-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the monoalkylated mitomycin C-DNA complex.
J.Mol.Biol., 247, 1995
|
|
