5TMM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC analog, (E)-6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6S)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMU
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 4,4'-(cycloheptylidenemethylene)diphenol | | Descriptor: | 4,4'-(cycloheptylidenemethylene)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.429 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TN4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the ACD-ring estrogen, (S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol | | Descriptor: | (1S)-5-(4-hydroxy-3,5-dimethylphenyl)-2,3-dihydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TLG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with (E)-4,4''-dihydroxy-3'-((hydroxyiminio)methyl)-[1,1':2',1''-terphenyl]-4'-olate | | Descriptor: | 2~3~-[(E)-(hydroxyimino)methyl][1~1~,2~1~:2~2~,3~1~-terphenyl]-1~4~,2~4~,3~4~-triol, Estrogen receptor, NUCLEAR RECEPTOR COACTIVATOR 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-11 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.228 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
1KAL
 
 | |
5TM8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
2C9Z
 
 | | Structure and activity of a flavonoid 3-0 glucosyltransferase reveals the basis for plant natural product modification | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, UDP GLUCOSE:FLAVONOID 3-O-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Offen, W, Martinez-Fleites, C, Kiat-Lim, E, Yang, M, Davis, B.G, Tarling, C.A, Ford, C.M, Bowles, D.J, Davies, G.J. | | Deposit date: | 2005-12-15 | | Release date: | 2006-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Flavonoid Glucosyltransferase Reveals the Basis for Plant Natural Product Modification.
Embo J., 25, 2006
|
|
2CCE
 
 | | Parallel Configuration of pLI E20S | | Descriptor: | General control protein GCN4 | | Authors: | Yadav, M.K, Leman, L.J, Price, D.J, Brooks 3rd, C.L, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2006-01-16 | | Release date: | 2006-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution.
Biochemistry, 45, 2006
|
|
2CHN
 
 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity- NAG-thiazoline complex | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
5WEZ
 
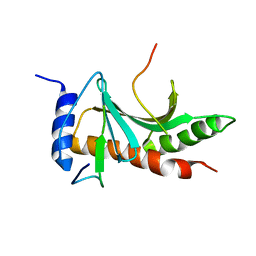 | | Structure of the Tir-CesT effector-chaperone complex | | Descriptor: | Tir chaperone, Translocated intimin receptor Tir | | Authors: | Little, D.J, Coombes, B.K. | | Deposit date: | 2017-07-11 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular basis for CesT recognition of type III secretion effectors in enteropathogenic Escherichia coli.
PLoS Pathog., 14, 2018
|
|
2CHO
 
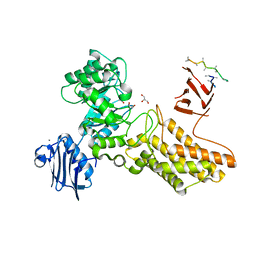 | | Bacteroides thetaiotaomicron hexosaminidase with O-GlcNAcase activity | | Descriptor: | ACETATE ION, CALCIUM ION, GLUCOSAMINIDASE, ... | | Authors: | Dennis, R.J, Taylor, E.J, Macauley, M.S, Stubbs, K.A, Turkenburg, J.P, Hart, S.J, Black, G.N, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2006-03-16 | | Release date: | 2006-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Mechanism of a Bacterial B-Glucosaminidase Having O-Glcnacase Activity
Nat.Struct.Mol.Biol., 13, 2006
|
|
5LY6
 
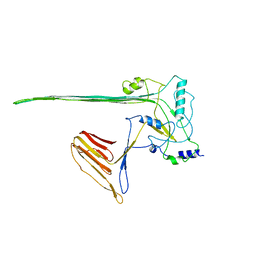 | | CryoEM structure of the membrane pore complex of Pneumolysin at 4.5A | | Descriptor: | Pneumolysin | | Authors: | van Pee, K, Neuhaus, A, D'Imprima, E, Mills, D.J, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2016-09-24 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin.
Elife, 6, 2017
|
|
5UDH
 
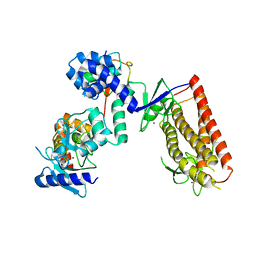 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
5UHK
 
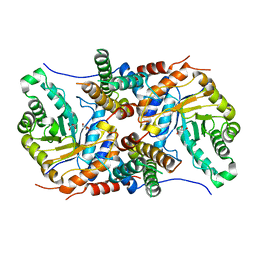 | |
5UHP
 
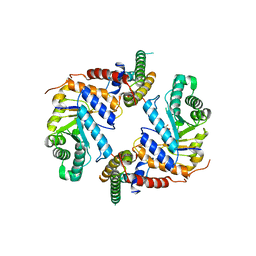 | |
2JZ4
 
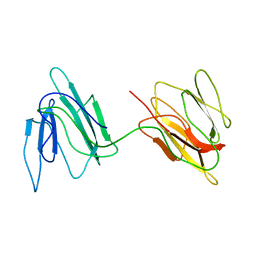 | | Putative 32 kDa myrosinase binding protein At3g16450.1 from Arabidopsis thaliana | | Descriptor: | Jasmonate inducible protein isolog | | Authors: | Takeda, N, Sugimori, N, Torizawa, T, Terauchi, T, Ono, A.M, Yagi, H, Yamaguchi, Y, Kato, K, Ikeya, T, Guntert, P, Aceti, D.J, Markley, J.L, Kainosho, M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the putative 32 kDa myrosinase-binding protein from Arabidopsis (At3g16450.1) determined by SAIL-NMR.
Febs J., 275, 2008
|
|
1OJM
 
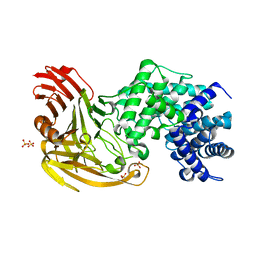 | |
1ONU
 
 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-G, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-G | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
2CAZ
 
 | | ESCRT-I core | | Descriptor: | PROTEIN SRN2, SUPPRESSOR PROTEIN STP22 OF TEMPERATURE-SENSITIVE ALPHA-FACTOR RECEPTOR AND ARGININE PERMEASE, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN VPS28 | | Authors: | Gill, D.J, Teo, H, Sun, J, Perisic, O, Veprintsev, D.B, Vallis, Y, Emr, S.D, Williams, R.L. | | Deposit date: | 2005-12-23 | | Release date: | 2006-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Escrt-I Core and Escrt-II Glue Domain Structures Reveal Role for Glue in Linking to Escrt-I and Membranes.
Cell(Cambridge,Mass.), 125, 2006
|
|
2CEX
 
 | | Structure of a sialic acid binding protein (SiaP) in the presence of the sialic acid acid analogue Neu5Ac2en | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, GLYCEROL, PROTEIN HI0146, ... | | Authors: | Muller, A, Severi, E, Mulligan, C, Watts, A.G, Kelly, D.J, Wilson, K.S, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2006-02-10 | | Release date: | 2006-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conservation of Structure and Mechanism in Primary and Secondary Transporters Exemplified by Siap, a Sialic Acid Binding Virulence Factor from Haemophilus Influenzae
J.Biol.Chem., 281, 2006
|
|
1OWC
 
 | | Three Dimensional Structure Analysis Of The R109L Variant of the Type II Citrate Synthase From E. Coli | | Descriptor: | Citrate synthase, SULFATE ION | | Authors: | Stokell, D.J, Donald, L.J, Maurus, R, Nguyen, N.T, Sadler, G, Choudhary, K, Hultin, P.G, Brayer, G.D, Duckworth, H.W. | | Deposit date: | 2003-03-28 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Probing the roles of key residues in the unique regulatory NADH binding site of type II citrate synthase of Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
2A90
 
 | |
243D
 
 | | STRUCTURE OF THE DNA OCTANUCLEOTIDE D(ACGTACGT)2 | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*T)-3') | | Authors: | Wilcock, D.J, Adams, A, Cardin, C.J, Wakelin, L.P.G. | | Deposit date: | 1996-01-10 | | Release date: | 1996-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the DNA octanucleotide d(ACGTACGT)2.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MFP
 
 | | E. coli Enoyl Reductase in complex with NAD and SB611113 | | Descriptor: | (E)-N-METHYL-N-(1-METHYL-1H-INDOL-3-YLMETHYL)-3-(7-OXO-5,6,7,8-TETRAHYDRO-[1,8]NAPHTHYRIDIN-3-YL)-ACRYLAMIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Seefeld, M.A, Miller, W.H, Newlander, K.A, Burgess, W.J, DeWolf Jr, W.E, Elkins, P.A, Head, M.S, Jakas, D.R, Janson, C.A, Keller, P.M, Manley, P.J, Moore, T.D, Payne, D.J, Pearson, S, Polizzi, B.J, Qiu, X, Rittenhouse, S.F, Uzinskas, I.N, Wallis, N.G, Huffman, W.F. | | Deposit date: | 2002-08-13 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Indole Naphthyridinones as Inhibitors of Bacterial Enoyl-ACP Reductases FabI and FabK
J.MED.CHEM., 46, 2003
|
|
2AB9
 
 | |
