4BLP
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI13 | | Descriptor: | CITRATE ANION, GLYCEROL, PACKAGING ENZYME P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4D6E
 
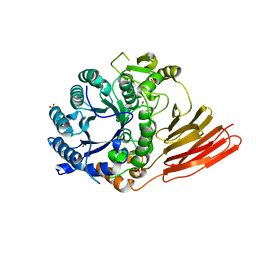 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the blood group A-trisaccharide (X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4MQL
 
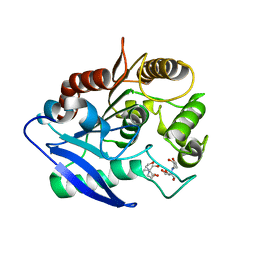 | | Crystal structure of Antigen 85C-C209S mutant | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
4D72
 
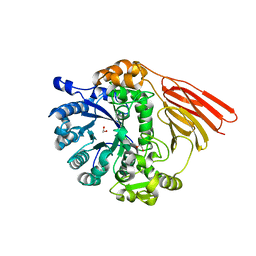 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 2 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCOSIDE HYDROLASE, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-19 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6F
 
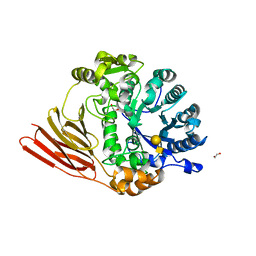 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A, X01 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6I
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
4D6H
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98) in complex with the type 1 blood group A-tetrasaccharide (E558A X02 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE, SULFATE ION, ... | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
6DZF
 
 | |
4QDT
 
 | |
5WEU
 
 | | Crystal Structure of H2-Dd with disulfide-linked 10mer peptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, Envelope glycoprotein gp160, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
4BTP
 
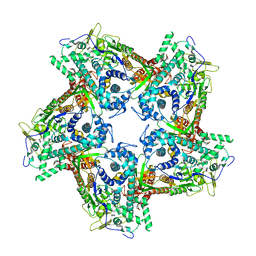 | | Structure of the capsid protein P1 of the bacteriophage phi8 | | Descriptor: | p1 | | Authors: | El Omari, K, Sutton, G, Ravantti, J.J, Zhang, H, Walter, T.S, Grimes, J.M, Bamford, D.H, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Plate Tectonics of Virus Shell Assembly and Reorganization in Phage Phi8, a Distant Relative of Mammalian Reoviruses
Structure, 21, 2013
|
|
5WET
 
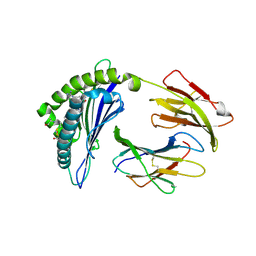 | | Crystal Structure of H2-Dd with disulfide-linked 6mer peptide | | Descriptor: | Beta-2-microglobulin, GLYCINE, H-2 class I histocompatibility antigen, ... | | Authors: | Jiang, J.S, Natarajan, K, Boyd, L.F, Margulies, D.H. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-18 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of a TAPBPR-MHC I complex reveals the mechanism of peptide editing in antigen presentation.
Science, 358, 2017
|
|
5WPR
 
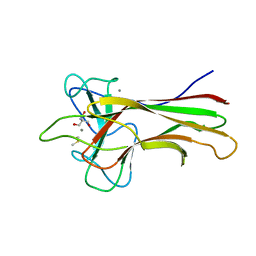 | | Crystal structure HpiC1 in C2 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5WPP
 
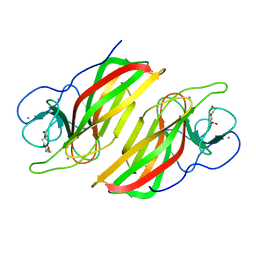 | | Crystal structure HpiC1 W73M/K132M | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
5XAC
 
 | | CLIR - LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5X9Q
 
 | | Crystal structure of HldC from Burkholderia pseudomallei | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of D-glycero-Beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei.
Proteins, 86, 2018
|
|
4MQM
 
 | | Crystal structure of Antigen 85C in presence of Ebselen | | Descriptor: | Diacylglycerol acyltransferase/mycolyltransferase Ag85C | | Authors: | Favrot, L, Grzegorzewicz, A.E, Lajiness, D.H, Marvin, R.K, Boucau, J, Isailovic, D, Jackson, M, Ronning, D.R. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Mechanism of inhibition of Mycobacterium tuberculosis antigen 85 by ebselen.
Nat Commun, 4, 2013
|
|
4D6C
 
 | | Crystal structure of a family 98 glycoside hydrolase catalytic module (Sp3GH98)(L19 mutant) | | Descriptor: | 1,2-ETHANEDIOL, GLYCOSIDE HYDROLASE | | Authors: | Kwan, D.H, Constantinescu, I, Chapanian, R, Higgins, M.A, Samain, E, Boraston, A.B, Kizhakkedathu, J.N, Withers, S.G. | | Deposit date: | 2014-11-11 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Towards Efficient Enzymes for the Generation of Universal Blood Through Structure-Guided Directed Evolution.
J.Am.Chem.Soc., 137, 2015
|
|
5X3T
 
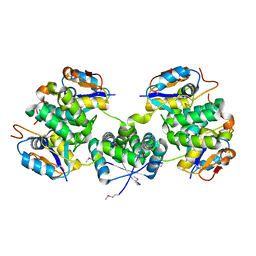 | | VapBC from Mycobacterium tuberculosis | | Descriptor: | Antitoxin VapB26, MAGNESIUM ION, Ribonuclease VapC26 | | Authors: | Kang, S.M, Kim, D.H, Yoon, H.J, Lee, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-07 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides.
Nucleic Acids Res., 45, 2017
|
|
5XAE
 
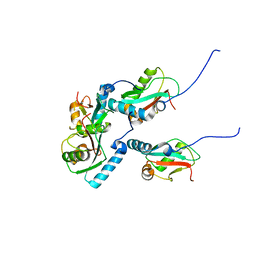 | | mutNLIR_LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5XF2
 
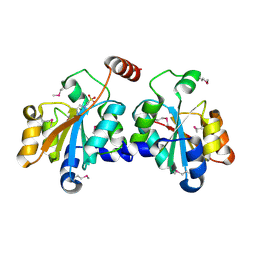 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4P6B
 
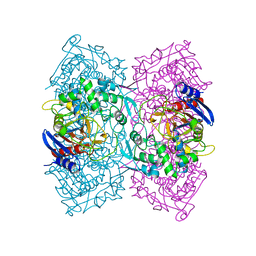 | | Crystal structure of Est-Y29,a novel penicillin-binding protein/beta-lactamase homolog from a metagenomic library | | Descriptor: | Est-Y29 | | Authors: | Ngo, T.D, Ryu, B.H, Ju, H.S, Jang, E.J, Kim, K.K, Kim, D.H. | | Deposit date: | 2014-03-24 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic analysis and biochemical applications of a novel penicillin-binding protein/ beta-lactamase homologue from a metagenomic library.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AW3
 
 | | Structure of the mixed-function P450 MycG F286V mutant in complex with mycinamicin V in P1 space group | | Descriptor: | GLYCEROL, MYCINAMICIN V, P-450-LIKE PROTEIN, ... | | Authors: | Li, S, Tietz, D.R, Rutaganira, F.U, Kells, P.M, Anzai, Y, Kato, F, Pochapsky, T.C, Sherman, D.H, Podust, L.M. | | Deposit date: | 2012-05-30 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
4BLT
 
 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S292A MUTANT IN COMPLEX WITH AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
4PR3
 
 | | Crystal structure of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Descriptor: | 5'-methylthioadenosine nucleosidase / s-adenosylhomocysteine nucleosidase, ADENINE, GLYCEROL, ... | | Authors: | Zhang, X.C, Kang, X.S, Zhao, Y, Jiang, D.H, Li, X.M, Chen, Z.L. | | Deposit date: | 2014-03-05 | | Release date: | 2014-04-30 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Crystal structure and biochemical studies of Brucella melitensis 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase
Biochem.Biophys.Res.Commun., 446, 2014
|
|
