1NTQ
 
 | | 5'(dCCUCCUU)3':3'(rAGGAGGAAA)5' | | Descriptor: | 5'-D(*CP*CP*UP*CP*CP*UP*U)-3', 5'-R(*AP*AP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Znosko, B.M, Barnes III, T.W, Krugh, T.R, Turner, D.H. | | Deposit date: | 2003-01-30 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Studies of DNA Single Strands and DNA:RNA Hybrids With and Without 1-Propynylation at C5 of Oligopyrimidines
J.Am.Chem.Soc., 125, 2003
|
|
1ODO
 
 | | 1.85 A structure of CYP154A1 from Streptomyces coelicolor A3(2) | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE, PUTATIVE CYTOCHROME P450 154A1 | | Authors: | Podust, L.M, Kim, Y, Arase, M, Bach, H, Sherman, D.H, Lamb, D.C, Kelly, S.L, Waterman, M.R. | | Deposit date: | 2003-02-19 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of the 1.85 A Structure of Cyp154A1 from Streptomyces Coelicolor A3(2) with the Closely Related Cyp154C1 and Cyps from Antibiotic Biosynthetic Pathways.
Protein Sci., 13, 2004
|
|
2B0Q
 
 | |
3R4Y
 
 | | Crystal structure of alpha-neoagarobiose hydrolase (ALPHA-NABH) from Saccharophagus degradans 2-40 | | Descriptor: | Glycosyl hydrolase family 32, N terminal | | Authors: | Lee, S, Lee, J.Y, Ha, S.C, Shin, D.H, Kim, K.H, Bang, W.G, Kim, S.H, Choi, I.G. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a key enzyme in the agarolytic pathway, alpha-neoagarobiose hydrolase from Saccharophagus degradans 2-40
Biochem.Biophys.Res.Commun., 412, 2011
|
|
2AXU
 
 | | Structure of PrgX | | Descriptor: | PrgX | | Authors: | Shi, K, Brown, C.K, Gu, Z.Y, Kozlowicz, B.K, Dunny, G.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2005-09-06 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1NP6
 
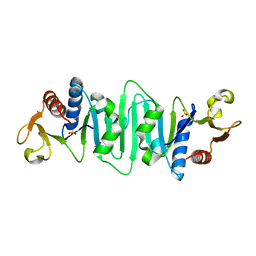 | | Crystal structure of Escherichia coli MobB | | Descriptor: | Molybdopterin-guanine dinucleotide biosynthesis protein B, SULFATE ION | | Authors: | McLuskey, K, Harrison, J.A, Schuttelkopf, A.W, Boxer, D.H, Hunter, W.N. | | Deposit date: | 2003-01-17 | | Release date: | 2003-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insight into the role of Escherichia coli MobB in Molybdenum cofactor biosynthesis based on the high resolution crystal structure
J.Biol.Chem., 278, 2003
|
|
1OIL
 
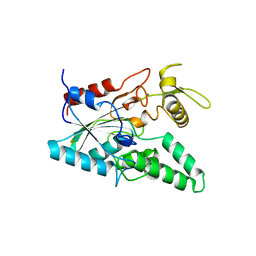 | | STRUCTURE OF LIPASE | | Descriptor: | CALCIUM ION, LIPASE | | Authors: | Kim, K.K, Song, H.K, Shin, D.H, Suh, S.W. | | Deposit date: | 1996-12-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a triacylglycerol lipase from Pseudomonas cepacia reveals a highly open conformation in the absence of a bound inhibitor.
Structure, 5, 1997
|
|
2CWB
 
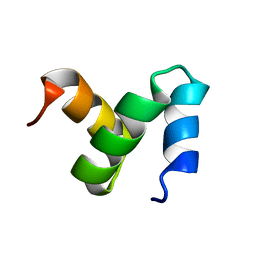 | | Solution Structure of the Ubiquitin-Associated Domain of Human BMSC-UbP and its Complex with Ubiquitin | | Descriptor: | Immunoglobulin G-binding protein G,Ubiquitin-like protein 7 | | Authors: | Chang, Y.G, Song, A.X, Gao, Y.G, Shi, Y.H, Lin, X.J, Cao, X.T, Lin, D.H, Hu, H.Y. | | Deposit date: | 2005-06-17 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin-associated domain of human BMSC-UbP and its complex with ubiquitin.
Protein Sci., 15, 2006
|
|
2BA2
 
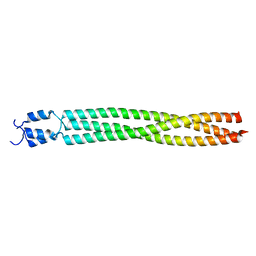 | | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae | | Descriptor: | Hypothetical UPF0134 protein MPN010 | | Authors: | Shin, D.H, Kim, J.-S, Yokota, H, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2005-10-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the DUF16 domain of MPN010 from Mycoplasma pneumoniae.
Protein Sci., 15, 2006
|
|
2ACQ
 
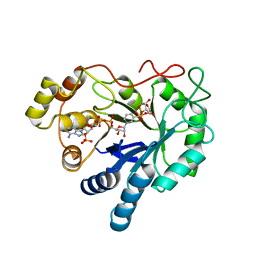 | | AN ANION BINDING SITE IN HUMAN ALDOSE REDUCTASE: MECHANISTIC IMPLICATIONS FOR THE BINDING OF CITRATE, CACODYLATE, AND GLUCOSE-6-PHOSPHATE | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H, Bohren, K.M, Gabbay, K.H, Petsko, G.A, Ringe, D. | | Deposit date: | 1994-04-15 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | An anion binding site in human aldose reductase: mechanistic implications for the binding of citrate, cacodylate, and glucose 6-phosphate.
Biochemistry, 33, 1994
|
|
2ADJ
 
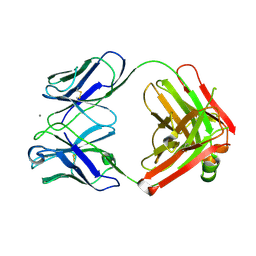 | | Crystal structure of monoclonal anti-CD4 antibody Q425 in complex with Calcium | | Descriptor: | CALCIUM ION, Q425 Fab Heavy chain, Q425 Fab Light chain | | Authors: | Zhou, T, Hamer, D.H, Hendrickson, W.A, Sattentau, Q.J, Kwong, P.D. | | Deposit date: | 2005-07-20 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interfacial metal and antibody recognition.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2C7X
 
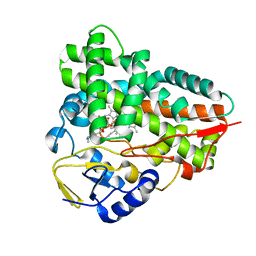 | | Crystal structure of narbomycin-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | CYTOCHROME P450 MONOOXYGENASE, NARBOMYCIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sherman, D.H, Li, S, Yermalitskaya, L.V, Kim, Y, Smith, J.A, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-11-29 | | Release date: | 2006-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2CA0
 
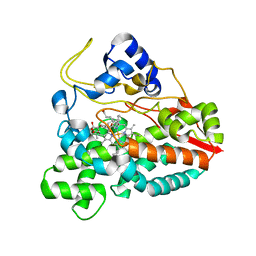 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Yc-17-Bound Cytochrome P450 Pikc (Cyp107L1)
To be Published
|
|
2CD8
 
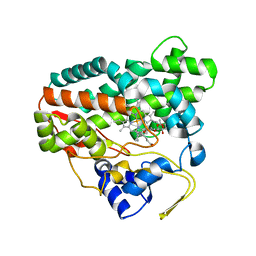 | | Crystal structure of YC-17-bound cytochrome P450 PikC (CYP107L1) | | Descriptor: | 4-{[4-(DIMETHYLAMINO)-3-HYDROXY-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL]OXY}-12-ETHYL-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yermalitskaya, L.I, Kim, Y, Sherman, D.H, Waterman, M.R, Podust, L.M. | | Deposit date: | 2006-01-20 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structural Basis for Substrate Anchoring, Active Site Selectivity, and Product Formation by P450 Pikc from Streptomyces Venezuelae.
J.Biol.Chem., 281, 2006
|
|
2K8C
 
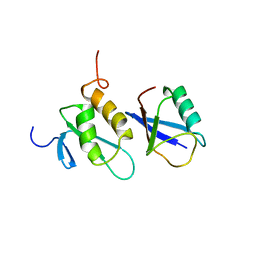 | | Solution structure of PLAA family ubiquitin binding domain (PFUC) trans isomer in complex with ubiquitin | | Descriptor: | Phospholipase A-2-activating protein, Ubiquitin | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2KY1
 
 | |
2KVJ
 
 | | NMR and MD solution structure of a Gamma-Methylated PNA duplex | | Descriptor: | Gamma-Modified Peptide Nucleic Acid | | Authors: | He, W, Crawford, M.J, Rapireddy, S, Madrid, M, Gil, R.R, Ly, D.H, Achim, C. | | Deposit date: | 2010-03-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of a gamma-modified peptide nucleic acid duplex.
Mol Biosyst, 6, 2010
|
|
2KY2
 
 | |
2KXZ
 
 | |
2LIW
 
 | | NMR structure of HMG-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, 4'-PHOSPHOPANTETHEINE, CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-21 | | Last modified: | 2012-03-14 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
1HRQ
 
 | | THE THREE-DIMENSIONAL SOLUTION STRUCTURE OF THE REDUCED HIGH-POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
1HRR
 
 | | THE THREE DIMENSIONAL STRUCTURE OF THE REDUCED HIGH POTENTIAL IRON-SULFUR PROTEIN FROM CHROMATIUM VINOSUM THROUGH NMR | | Descriptor: | IRON/SULFUR CLUSTER, REDUCED HIGH POTENTIAL IRON SULFUR PROTEIN | | Authors: | Banci, L, Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | Deposit date: | 1995-01-17 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional solution structure of the reduced high-potential iron-sulfur protein from Chromatium vinosum through NMR.
Biochemistry, 34, 1995
|
|
2JLS
 
 | | Dengue virus 4 NS3 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLU
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA | | Descriptor: | 5'-R(*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', GLYCEROL, SERINE PROTEASE SUBUNIT NS3 | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein.
Embo J., 27, 2008
|
|
2JLW
 
 | | Dengue virus 4 NS3 helicase in complex with ssRNA2 | | Descriptor: | 5'-R(*UP*AP*GP*AP*CP*UP*AP*AP*CP*AP*AP*CP*U)-3', GLYCEROL, PHOSPHATE ION, ... | | Authors: | Luo, D.H, Xu, T, Watson, R.P, Becker, D.S, Sampath, A, Jahnke, W, Yeong, S.S, Wang, C.H, Lim, S.P, Vasudevan, S.G, Lescar, J. | | Deposit date: | 2008-09-15 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into RNA Unwinding and ATP Hydrolysis by the Flavivirus Ns3 Protein
Embo J., 27, 2008
|
|
