6R8I
 
 | | PP4R3A EVH1 domain bound to FxxP motif | | Descriptor: | SER-LEU-PRO-PHE-THR-PHE-LYS-VAL-PRO-ALA-PRO-PRO-PRO-SER-LEU-PRO-PRO-SER, Serine/threonine-protein phosphatase 4 regulatory subunit 3A | | Authors: | Ueki, Y, Kruse, T, Weisser, M.B, Sundell, G.N, Yoo Larsen, M.S, Lopez Mendez, B, Jenkins, N.P, Garvanska, D.H, Cressey, L, Zhang, G, Davey, N, Montoya, G, Ivarsson, Y, Kettenbach, A, Nilsson, J. | | Deposit date: | 2019-04-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.517 Å) | | Cite: | A Consensus Binding Motif for the PP4 Protein Phosphatase.
Mol.Cell, 76, 2019
|
|
7S3T
 
 | | NzeB Diketopiperazine Dimerase Mutant: Q68I-G87A-A89G-I90V | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, 1,2-ETHANEDIOL, MAGNESIUM ION, ... | | Authors: | Harris, N.R, Shende, V.V, Sanders, J.N, Newmister, S.A, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-08 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 2023
|
|
5X9Q
 
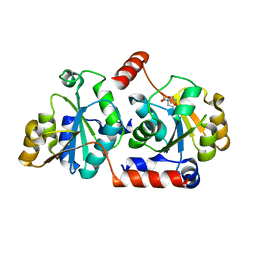 | | Crystal structure of HldC from Burkholderia pseudomallei | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-03-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of D-glycero-Beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei.
Proteins, 86, 2018
|
|
5XAC
 
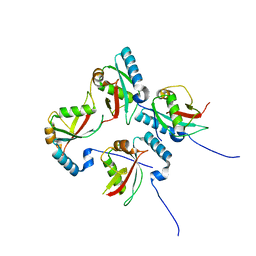 | | CLIR - LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
7S3J
 
 | | Crystal Structure of AspB P450 in complex with brevianamide F substrates | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, AspB, GLYCEROL, ... | | Authors: | Newmister, S.A, Shende, V.V, Harris, N.R, Sanders, J.N, Khatri, Y, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2021-09-07 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Dynamics Simulations Guide Chimeragenesis and Engineered Control of Chemoselectivity in Diketopiperazine Dimerases.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5XF2
 
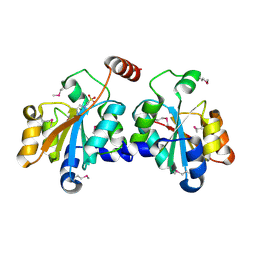 | | Crystal structure of SeMet-HldC from Burkholderia pseudomallei | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative cytidylyltransferase | | Authors: | Park, J, Kim, H, Kim, S, Lee, D, Shin, D.H. | | Deposit date: | 2017-04-07 | | Release date: | 2017-07-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Expression and crystallographic studies of D-glycero-beta-D-manno-heptose-1-phosphate adenylyltransferase from Burkholderia pseudomallei
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5XAE
 
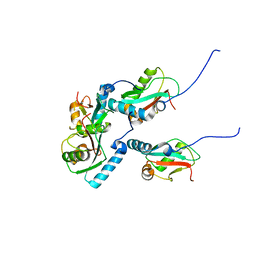 | | mutNLIR_LC3B | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
5X3T
 
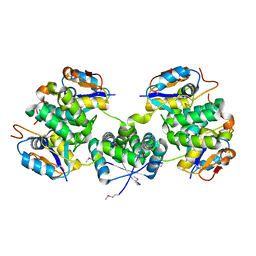 | | VapBC from Mycobacterium tuberculosis | | Descriptor: | Antitoxin VapB26, MAGNESIUM ION, Ribonuclease VapC26 | | Authors: | Kang, S.M, Kim, D.H, Yoon, H.J, Lee, B.J. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-07 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional details of the Mycobacterium tuberculosis VapBC26 toxin-antitoxin system based on a structural study: insights into unique binding and antibiotic peptides.
Nucleic Acids Res., 45, 2017
|
|
6RZN
 
 | |
1HHS
 
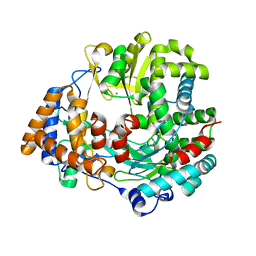 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
5XAD
 
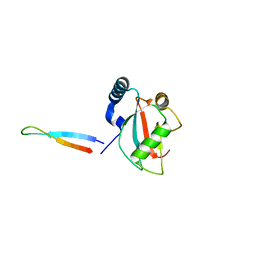 | | NLIR - LC3B fusion protein | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Uncharacterised protein | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
1HI0
 
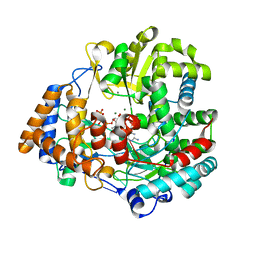 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus initiation complex | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-31 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
6RZO
 
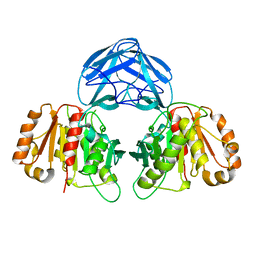 | |
1IUC
 
 | | Fucose-specific lectin from Aleuria aurantia with three ligands | | Descriptor: | Fucose-specific lectin, SULFATE ION, alpha-L-fucopyranose, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
6SU7
 
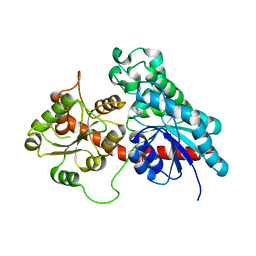 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and 3,4-Dichloroaniline | | Descriptor: | 3,4-Dichloroaniline, Glycosyltransferase | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
6SU6
 
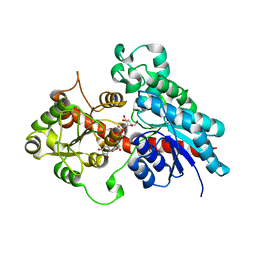 | | Complex between a UDP-glucosyltransferase from Polygonum tinctorium capable of glucosylating indoxyl and UDP-glucose | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Fredslund, F, Teze, D, Svensson, B, Adams, P.D, Welner, D.H. | | Deposit date: | 2019-09-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | O-/N-/S-Specificity in Glycosyltransferase Catalysis: From Mechanistic Understanding to Engineering
Acs Catalysis, 11, 2021
|
|
6T2B
 
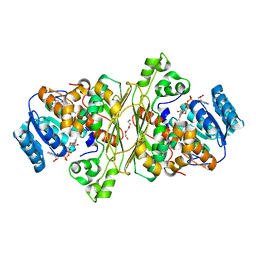 | | Glycoside hydrolase family 109 from Akkermansia muciniphila in complex with GalNAc and NAD+. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Glycosyl hydrolase family 109 protein 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chaberski, E.K, Fredslund, F, Teze, D, Shuoker, B, Kunstmann, S, Karlsson, E.N, Hachem, M.A, Welner, D.H. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | The Catalytic Acid-Base in GH109 Resides in a Conserved GGHGG Loop and Allows for Comparable alpha-Retaining and beta-Inverting Activity in an N-Acetylgalactosaminidase from Akkermansia muciniphila
Acs Catalysis, 2020
|
|
1HHT
 
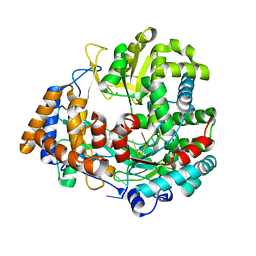 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus template | | Descriptor: | DNA (5'-(*TP*TP*TP*CP*C)-3'), MANGANESE (II) ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2000-12-28 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1HI1
 
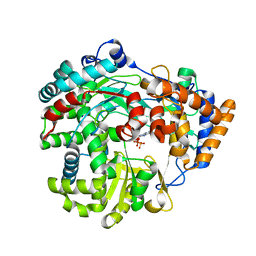 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 plus bound NTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-01 | | Release date: | 2001-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1HI8
 
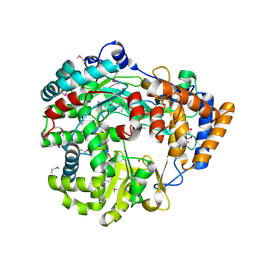 | | RNA dependent RNA polymerase from dsRNA bacteriophage phi6 | | Descriptor: | MAGNESIUM ION, P2 PROTEIN | | Authors: | Grimes, J.M, Butcher, S.J, Makeyev, E.V, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2001-01-03 | | Release date: | 2001-03-27 | | Last modified: | 2019-09-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Mechanism for Initiating RNA-Dependent RNA Polymerization
Nature, 410, 2001
|
|
1IUB
 
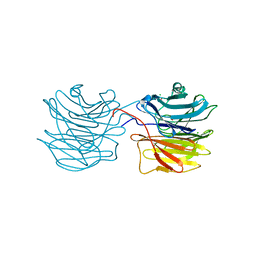 | | Fucose-specific lectin from Aleuria aurantia (Hg-derivative form) | | Descriptor: | CHLORIDE ION, Fucose-specific lectin, MERCURY (II) ION, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
1JA3
 
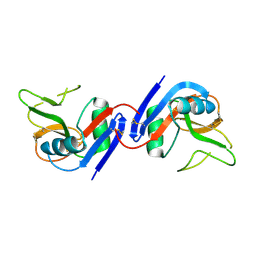 | | Crystal Structure of the Murine NK Cell Inhibitory Receptor Ly-49I | | Descriptor: | MHC class I recognition receptor Ly49I | | Authors: | Dimasi, N, Sawicki, W.M, Reineck, L.A, Li, Y, Natarajan, K, Murgulies, D.H, Mariuzza, A.R. | | Deposit date: | 2001-05-29 | | Release date: | 2002-07-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Ly49I natural killer cell receptor reveals variability in dimerization mode within the Ly49 family.
J.Mol.Biol., 320, 2002
|
|
5IZV
 
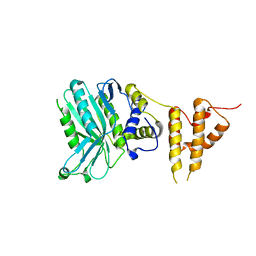 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
7YK6
 
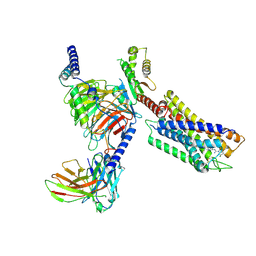 | | Cryo-EM structure of the compound 4-bound human relaxin family peptide receptor 4 (RXFP4)-Gi complex | | Descriptor: | 1-[2-(4-chlorophenyl)ethyl]-3-[(7-ethyl-5-oxidanyl-1H-indol-3-yl)methylideneamino]guanidine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, Y, Zhou, Q.T, Wang, J, Xu, Y.W, Wang, Y, Yan, J.H, Wang, Y.B, Zhu, Q, Zhao, F.H, Li, C.H, Chen, C.W, Cai, X.Q, Bathgate, R.A.D, Shen, C, Liu, H, Xu, H.E, Yang, D.H, Wang, M.W. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Ligand recognition mechanism of the human relaxin family peptide receptor 4 (RXFP4).
Nat Commun, 14, 2023
|
|
7O6H
 
 | |
