2YIA
 
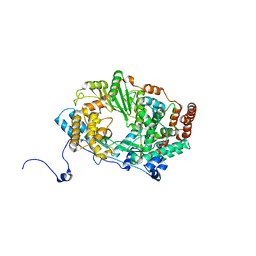 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2YM4
 
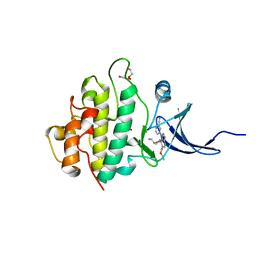 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1, ethyl 4-[(2R)-2-(aminomethyl)morpholin-4-yl]-3-(3-cyanophenyl)-1H-pyrazolo[3,4-b]pyridine-5-carboxylate | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2YI8
 
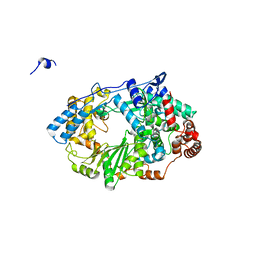 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | Descriptor: | CHLORIDE ION, POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | Deposit date: | 2011-05-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
6B4G
 
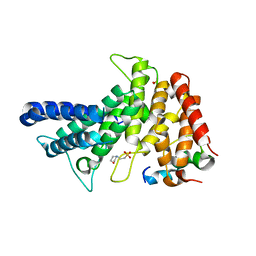 | | Crystal structure of Chaetomium thermophilum Gle1 CTD-Nup42 GBM complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoporin AMO1, Nucleoporin GLE1 | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
2YM5
 
 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (3-{4-[(2S)-2-(AMINOMETHYL)MORPHOLIN-4-YL]-7H-PYRROLO[2,3-D]PYRIMIDIN-5-YL}PHENYL)METHANOL, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
4A8Y
 
 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*TP*TP*CP*GP*CP*GP*TP*AP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
6B4I
 
 | | Crystal structure of human Gle1 CTD-Nup42 GBM-DDX19B(ADP) complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX19B, Nucleoporin GLE1, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.619 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
6B6P
 
 | |
2YM8
 
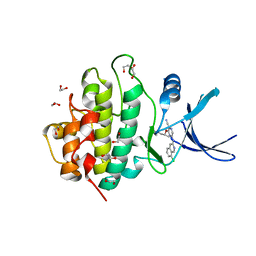 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | (R)-5-(8-CHLOROISOQUINOLIN-3-YLAMINO)-3-(1-(DIMETHYLAMINO)PROPAN-2-YLOXY)PYRAZINE-2-CARBONITRILE, 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Reader, J.C, Matthews, T.P, Klair, S, Cheung, K.M.J, Scanlon, J, Proisy, N, Addison, G, Ellard, J, Piton, N, Taylor, S, Cherry, M, Fisher, M, Boxall, K, Burns, S, Walton, M.I, Westwood, I.M, Hayes, A, Eve, P, Valenti, M, Brandon, A.H, Box, G, vanMontfort, R.L.M, Williams, D.H, Aherne, G.W, Raynaud, F.I, Eccles, S.A, Garrett, M.D, Collins, I. | | Deposit date: | 2011-06-06 | | Release date: | 2012-01-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Evolution of Potent and Selective Chk1 Inhibitors Through Scaffold Morphing.
J.Med.Chem., 54, 2011
|
|
2Y3S
 
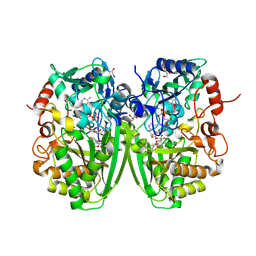 | | Structure of the tirandamycine-bound FAD-dependent tirandamycin oxidase TamL in C2 space group | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-12-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
5Z0B
 
 | |
2Y08
 
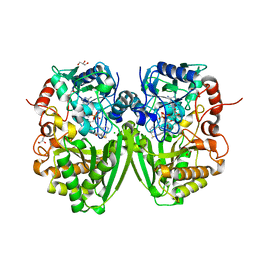 | | Structure of the substrate-free FAD-dependent tirandamycin oxidase TamL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Carlson, J.C, Li, S, Gunatilleke, S.S, Anzai, Y, Burr, D.A, Podust, L.M, Sherman, D.H. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tirandamycin Biosynthesis is Mediated by Co-Dependent Oxidative Enzymes
Nat.Chem, 3, 2011
|
|
2Y46
 
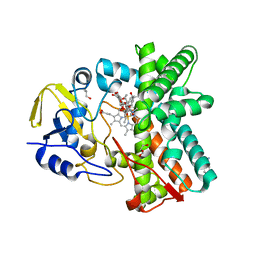 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in C 2 2 21 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y5N
 
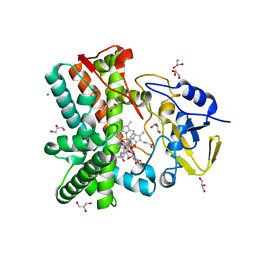 | | Structure of the mixed-function P450 MycG in complex with mycinamicin V in P21 space group | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYCINAMICIN V, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
4A8W
 
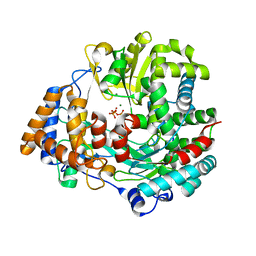 | | Non-Catalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial dsRNA virus phi6 from De Novo Initiation to Elongation | | Descriptor: | 5'-D(*TP*TP*CP*GP*CP*GP*TP*AP*AP*GP*CP*GP)-3', ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wright, S, Poranen, M.M, Bamford, D.H, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Noncatalytic Ions Direct the RNA-Dependent RNA Polymerase of Bacterial Double-Stranded RNA Virus Phi6 from De Novo Initiation to Elongation.
J.Virol., 86, 2012
|
|
2WHW
 
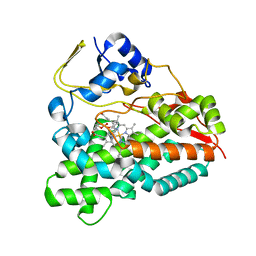 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLOTRIDECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 MONOOXYGENASE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-07 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6D87
 
 | |
2WI9
 
 | | Selective oxidation of carbolide C-H bonds by engineered macrolide P450 monooxygenase | | Descriptor: | CYCLODODECYL 3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSIDE, CYTOCHROME P450 HYDROXYLASE PIKC, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Li, S, Chaulagain, M.R, Knauff, A.R, Podust, L.M, Montgomery, J, Sherman, D.H. | | Deposit date: | 2009-05-08 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective Oxidation of Carbolide C-H Bonds by an Engineered Macrolide P450 Mono-Oxygenase.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6E47
 
 | |
6E48
 
 | | Crystal Structure of the Murine Norovirus VP1 P domain in complex with the CD300lf Receptor and Lithocholic Acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Nelson, C.A, Fremont, D.H. | | Deposit date: | 2018-07-16 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4BTP
 
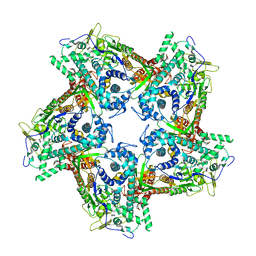 | | Structure of the capsid protein P1 of the bacteriophage phi8 | | Descriptor: | p1 | | Authors: | El Omari, K, Sutton, G, Ravantti, J.J, Zhang, H, Walter, T.S, Grimes, J.M, Bamford, D.H, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Plate Tectonics of Virus Shell Assembly and Reorganization in Phage Phi8, a Distant Relative of Mammalian Reoviruses
Structure, 21, 2013
|
|
4BLS
 
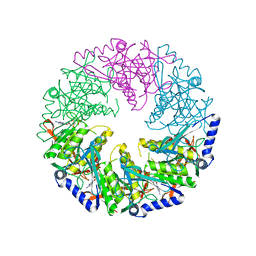 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 Q278A MUTANT IN COMPLEX WITH AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | El Omari, K, Meier, C, Kainov, D, Sutton, G, Grimes, J.M, Poranen, M.M, Bamford, D.H, Tuma, R, Stuart, D.I, Mancini, E.J. | | Deposit date: | 2013-05-04 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Tracking in Atomic Detail the Functional Specializations in Viral Reca Helicases that Occur During Evolution.
Nucleic Acids Res., 41, 2013
|
|
6B6R
 
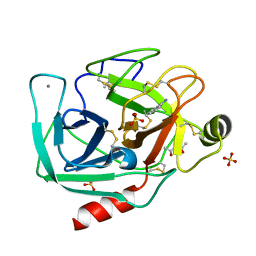 | | Orthorhombic trypsin cryocooled to 100 K with 50% mpd as cryoprotectant | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Juers, D.H. | | Deposit date: | 2017-10-03 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.00005865 Å) | | Cite: | The impact of cryosolution thermal contraction on proteins and protein crystals: volumes, conformation and order.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6B6Q
 
 | |
6B4F
 
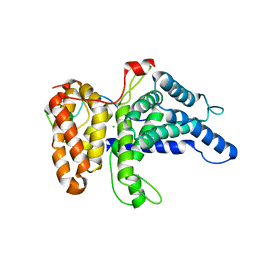 | | Crystal structure of human Gle1 CTD-Nup42 GBM complex | | Descriptor: | CHLORIDE ION, Nucleoporin GLE1, Nucleoporin like 2, ... | | Authors: | Lin, D.H, Correia, A.R, Cai, S.W, Huber, F.M, Jette, C.A, Hoelz, A. | | Deposit date: | 2017-09-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.811 Å) | | Cite: | Structural and functional analysis of mRNA export regulation by the nuclear pore complex.
Nat Commun, 9, 2018
|
|
