2BXJ
 
 | | Double Mutant of the Ribosomal Protein S6 | | Descriptor: | 30S RIBOSOMAL PROTEIN S6 | | Authors: | Otzen, D.E. | | Deposit date: | 2005-07-26 | | Release date: | 2005-10-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antagonism, Non-Native Interactions and Non-Two-State Folding in S6 Revealed by Double-Mutant Cycle Analysis.
Protein Eng.Des.Sel., 18, 2005
|
|
204D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
203D
 
 | | THE SOLUTION STRUCTURES OF PSORALEN MONOADDUCTED AND CROSSLINKED DNA OLIGOMERS BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | 4'-HYDROXYMETHYL-4,5',8-TRIMETHYLPSORALEN, DNA (5'-D(*GP*CP*GP*TP*AP*CP*GP*C)-3') | | Authors: | Spielmann, H.P, Dwyer, T.J, Hearst, J.E, Wemmer, D.E. | | Deposit date: | 1995-04-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 34, 1995
|
|
2BBI
 
 | |
2B0M
 
 | | Human dihydroorotate dehydrogenase bound to a novel inhibitor | | Descriptor: | 3-AMIDO-5-BIPHENYL-BENZOIC ACID, Dihydroorotate dehydrogenase, mitochondrial, ... | | Authors: | Hurt, D.E, Sutton, A.E, Clardy, J. | | Deposit date: | 2005-09-14 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Brequinar derivatives and species-specific drug design for dihydroorotate dehydrogenase.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2QS2
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
1CMT
 
 | | THE ROLE OF ASPARTATE-235 IN THE BINDING OF CATIONS TO AN ARTIFICIAL CAVITY AT THE RADICAL SITE OF CYTOCHROME C PEROXIDASE | | Descriptor: | CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fitzgerald, M.M, Trester, M.L, Jensen, G.M, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1995-04-11 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The role of aspartate-235 in the binding of cations to an artificial cavity at the radical site of cytochrome c peroxidase.
Protein Sci., 4, 1995
|
|
1FOX
 
 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, 33 STRUCTURES | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1FOW
 
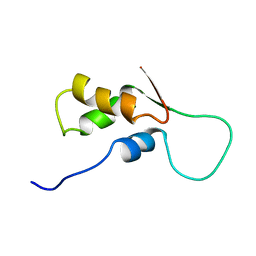 | | NMR STRUCTURE OF L11-C76, THE C-TERMINAL DOMAIN OF 50S RIBOSOMAL PROTEIN L11, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | L11-C76 | | Authors: | Markus, M.A, Hinck, A.P, Huang, S, Draper, D.E, Torchia, D.A. | | Deposit date: | 1996-09-13 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High resolution solution structure of ribosomal protein L11-C76, a helical protein with a flexible loop that becomes structured upon binding to RNA.
Nat.Struct.Biol., 4, 1997
|
|
1F4V
 
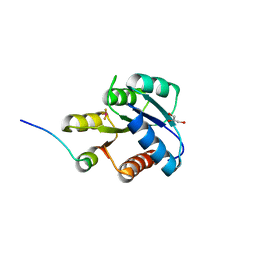 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
2QS4
 
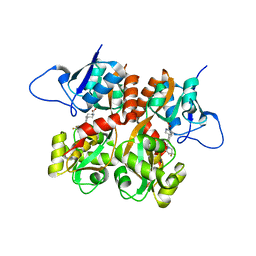 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
1S6N
 
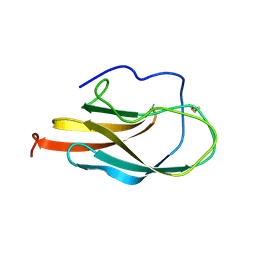 | | NMR Structure of Domain III of the West Nile Virus Envelope Protein, Strain 385-99 | | Descriptor: | envelope glycoprotein | | Authors: | Volk, D.E, Beasley, D.W, Kallick, D.A, Holbrook, M.R, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2004-01-26 | | Release date: | 2004-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Antibody Binding Studies of the Envelope Protein Domain III from the New York Strain of West Nile Virus
J.Biol.Chem., 279, 2004
|
|
4N9R
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentacholrocarbonyliridate(III) (1 day) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-10-21 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHQ
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (5 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHP
 
 | | X-ray structure of the complex between the hen egg white lysozyme and pentachlorocarbonyliridate (III) (4 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHT
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (6 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NHS
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (9 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-05 | | Release date: | 2014-09-17 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NIJ
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (30 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-09-17 | | Last modified: | 2015-06-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
1MT1
 
 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannaschii | | Descriptor: | AGMATINE, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE ALPHA CHAIN, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE BETA CHAIN | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1O0S
 
 | | Crystal Structure of Ascaris suum Malic Enzyme Complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NAD-dependent malic enzyme, TARTRONATE | | Authors: | Rao, G.S, Coleman, D.E, Karsten, W.E, Cook, P.F, Harris, B.G. | | Deposit date: | 2003-02-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site.
J.Biol.Chem., 278, 2003
|
|
1OY6
 
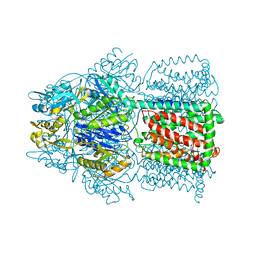 | | Structural Basis of the Multiple Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.68 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1OY8
 
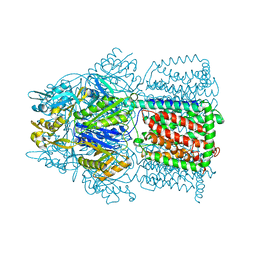 | | Structural Basis of Multiple Drug Binding Capacity of the AcrB Multidrug Efflux Pump | | Descriptor: | Acriflavine resistance protein B, RHODAMINE 6G | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
1PB3
 
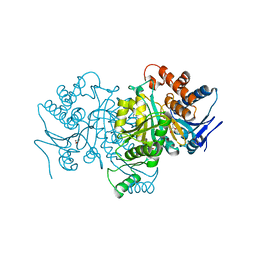 | |
1PB1
 
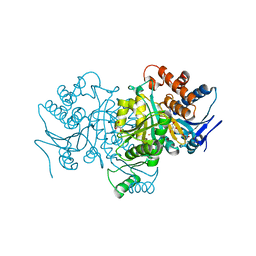 | |
1OYE
 
 | | Structural Basis of Multiple Binding Capacity of the AcrB multidrug Efflux Pump | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, Acriflavine resistance protein B | | Authors: | Yu, E.W, McDermott, G, Zgurskaya, H.I, Nikaido, H, Koshland Jr, D.E. | | Deposit date: | 2003-04-03 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump.
Science, 300, 2003
|
|
