1TM3
 
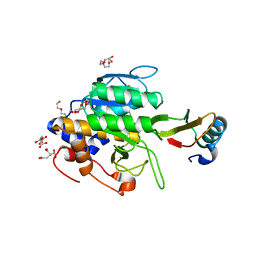 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59k mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-10 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
1SXL
 
 | |
1PQQ
 
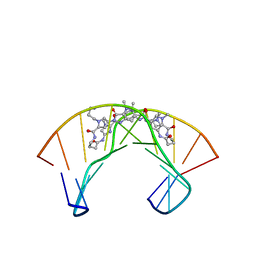 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
1Z3N
 
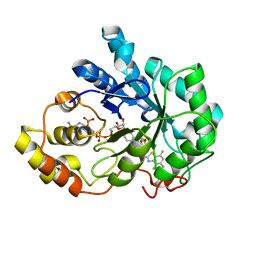 | | Human aldose reductase in complex with NADP+ and the inhibitor lidorestat at 1.04 angstrom | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase, {3-[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]-1H-INDOL-1-YL}ACETIC ACID | | Authors: | Van Zandt, M.C, Jones, M.L, Gunn, D.E, Geraci, L.S, Jones, J.H, Sawicki, D.R, Sredy, J, Jacot, J.L, Dicioccio, A.T, Petrova, T, Mitschler, A, Podjarny, A.D. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Discovery of 3-[(4,5,7-trifluorobenzothiazol-2-yl)methyl]indole-N-acetic acid (lidorestat) and congeners as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications
J.Med.Chem., 48, 2005
|
|
1PS7
 
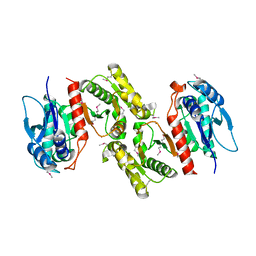 | | Crystal structure of E.coli PdxA | | Descriptor: | 4-hydroxythreonine-4-phosphate dehydrogenase, ZINC ION | | Authors: | Sivaraman, J, Li, Y, Banks, J, Cane, D.E, Matte, A, Cygler, M. | | Deposit date: | 2003-06-20 | | Release date: | 2003-11-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of Escherichia coli PdxA, an Enzyme Involved in the Pyridoxal Phosphate Biosynthesis Pathway
J.Biol.Chem., 278, 2003
|
|
1TO1
 
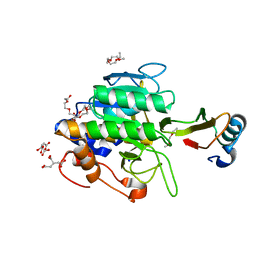 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 Y61A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
1Q35
 
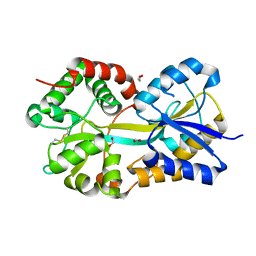 | | Crystal Structure of Pasteurella haemolytica Apo Ferric ion-Binding Protein A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, iron binding protein FbpA | | Authors: | Shouldice, S.R, Dougan, D.R, Skene, R.J, Snell, G, Scheibe, D, Williams, P.A, Kirby, S, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2003-07-28 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of Pasteurella haemolytica ferric ion-binding protein A reveals a novel class of bacterial iron-binding proteins
J.Biol.Chem., 278, 2003
|
|
1QCN
 
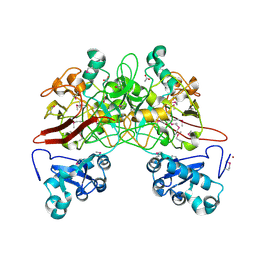 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE | | Descriptor: | ACETATE ION, CALCIUM ION, FUMARYLACETOACETATE HYDROLASE, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-14 | | Release date: | 2000-06-07 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1UN4
 
 | |
1QQJ
 
 | | CRYSTAL STRUCTURE OF MOUSE FUMARYLACETOACETATE HYDROLASE REFINED AT 1.55 ANGSTROM RESOLUTION | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-06-07 | | Release date: | 2000-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
1DSO
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX AT PH 6, ROOM TEMPERATURE. | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1DSE
 
 | | CYTOCHROME C PEROXIDASE H175G MUTANT, IMIDAZOLE COMPLEX, WITH PHOSPHATE BOUND, PH 6, 100K | | Descriptor: | CYTOCHROME C PEROXIDASE, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Hirst, J, Wilcox, S.K, Williams, P.A, McRee, D.E, Goodin, D.B. | | Deposit date: | 2000-01-07 | | Release date: | 2001-03-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Replacement of the axial histidine ligand with imidazole in cytochrome c peroxidase. 1. Effects on structure.
Biochemistry, 40, 2001
|
|
1SI0
 
 | | Crystal Structure of Mannheimia haemolytica Ferric iron-Binding Protein A in a closed conformation | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, Snell, G, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens.
J.Bacteriol., 186, 2004
|
|
1DK6
 
 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1SF9
 
 | | Crystal Structure of Bacillus subtilis YfhH Protein : Putative Transcriptional Regulator | | Descriptor: | CHLORIDE ION, PLATINUM (II) ION, yfhH hypothetical protein | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kim, D.E, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-19 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structure of Bacillus Subtilis YfhH hypothetical protein
To be Published
|
|
1YYQ
 
 | | Y305F Trichodiene Synthase complexed with pyrophosphate | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Trichodiene synthase | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1NP4
 
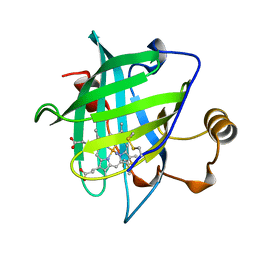 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS | | Descriptor: | AMMONIA, PROTEIN (NITROPHORIN 4), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Andersen, J.F, Weichsel, A, Champagne, D.E, Balfour, C.A, Montfort, W.R. | | Deposit date: | 1998-07-29 | | Release date: | 1998-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of nitrophorin 4 at 1.5 A resolution: transport of nitric oxide by a lipocalin-based heme protein.
Structure, 6, 1998
|
|
1QCI
 
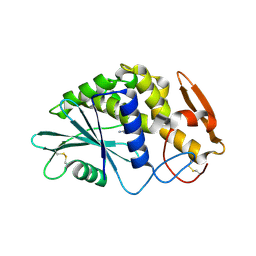 | | LOW TEMPERATURE STRUCTURE OF POKEWEED ANTIVIRAL PROTEIN COMPLEXED WITH ADENINE | | Descriptor: | ADENINE, POKEWEED ANTIVIRAL PROTEIN | | Authors: | Kurinov, I.V, Myers, D.E, Irvin, J.D, Uckun, F.M. | | Deposit date: | 1999-05-05 | | Release date: | 1999-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic analysis of the structural basis for the interactions of pokeweed antiviral protein with its active site inhibitor and ribosomal RNA substrate analogs.
Protein Sci., 8, 1999
|
|
1QWG
 
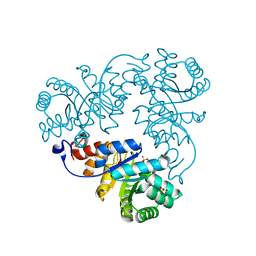 | | Crystal structure of Methanococcus jannaschii phosphosulfolactate synthase | | Descriptor: | (2R)-phospho-3-sulfolactate synthase, SULFATE ION | | Authors: | Wise, E.L, Graham, D.E, White, R.H, Rayment, I. | | Deposit date: | 2003-09-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structural determination of phosphosulfolactate synthase from Methanococcus jannaschii at 1.7-A resolution: an enolase that is not an enolase
J.Biol.Chem., 278, 2003
|
|
1UZI
 
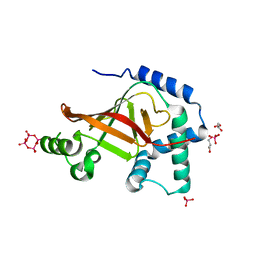 | | C3 EXOENZYME FROM CLOSTRIDIUM BOTULINUM, TETRAGONAL FORM | | Descriptor: | CYCLO-TETRAMETAVANADATE, GLYCEROL, MONO-ADP-RIBOSYLTRANSFERASE C3, ... | | Authors: | Evans, H.R, Holloway, D.E, Sutton, J.M, Ayriss, J, Shone, C.C, Acharya, K.R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | C3 Exoenzyme from Clostridium Botulinum: Structure of a Tetragonal Crystal Form and a Reassessment of Nad-Induced Flexure
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1N3L
 
 | | Crystal structure of a human aminoacyl-tRNA synthetase cytokine | | Descriptor: | GLYCEROL, SULFATE ION, tyrosyl-tRNA synthetase | | Authors: | Yang, X.-L, Skene, R.J, McRee, D.E, Schimmel, P. | | Deposit date: | 2002-10-28 | | Release date: | 2002-11-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Crystal structure of a human aminoacyl-tRNA synthetase cytokine
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1TBH
 
 | | H141D mutant of rat liver arginase I | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-20 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
1TM4
 
 | | crystal structure of the complex of subtilsin BPN'with chymotrypsin inhibitor 2 M59G mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-10 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
1MZD
 
 | | crystal structure of human pro-granzyme K | | Descriptor: | pro-granzyme K | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Wilharm, E, Fuentes-Prior, P, Klinkert, W, Bode, W, Jenne, D.E. | | Deposit date: | 2002-10-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.2-A Crystal Structure of Human Pro-granzyme K Reveals a Rigid Zymogen with Unusual Features
J.BIOL.CHEM., 277, 2002
|
|
1TM9
 
 | | NMR Structure of gene target number gi3844938 from Mycoplasma genitalium: Berkeley Structural Genomics Center | | Descriptor: | Hypothetical protein MG354 | | Authors: | Pelton, J.G, Shi, J, Yokota, H, Kim, R, Wemmer, D.E, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-06-10 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Gene Target gi3844938 from Mycoplasma genitalium
To be Published
|
|
