4QNH
 
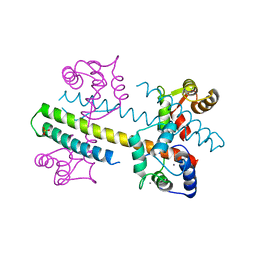 | | Calcium-calmodulin (T79D) complexed with the calmodulin binding domain from a small conductance potassium channel SK2-a | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Zhang, M, Pascal, J.M, Logothetis, D.E, Zhang, J.F. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Selective phosphorylation modulates the PIP2 sensitivity of the CaM-SK channel complex.
Nat.Chem.Biol., 10, 2014
|
|
1P8N
 
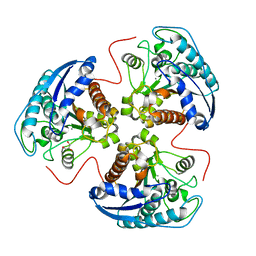 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Manganese Cluster of Arginase I. | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1OJD
 
 | |
1P31
 
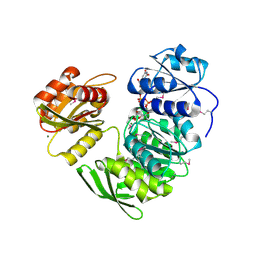 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine Ligase (MurC) from Haemophilus influenzae | | Descriptor: | MAGNESIUM ION, UDP-N-acetylmuramate--alanine ligase, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-16 | | Release date: | 2003-07-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
7Q58
 
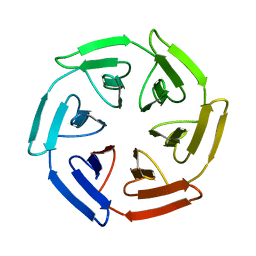 | | Crystal structure of the SAKe6BR designer protein | | Descriptor: | SAKe6BR | | Authors: | Wouters, S.M.L, Noguchi, H, Velupla, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-11-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
1P3D
 
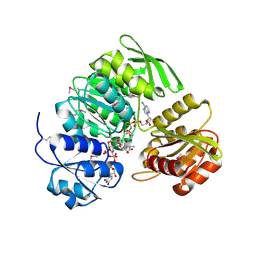 | | Crystal Structure of UDP-N-acetylmuramic acid:L-alanine ligase (MurC) in Complex with UMA and ANP. | | Descriptor: | MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, UDP-N-acetylmuramate--alanine ligase, ... | | Authors: | Mol, C.D, Brooun, A, Dougan, D.R, Hilgers, M.T, Tari, L.W, Wijnands, R.A, Knuth, M.W, McRee, D.E, Swanson, R.V. | | Deposit date: | 2003-04-17 | | Release date: | 2003-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structures of Active Fully Assembled Substrate- and Product-Bound Complexes of UDP-N-Acetylmuramic Acid:L-Alanine Ligase (MurC) from Haemophilus influenzae.
J.Bacteriol., 185, 2003
|
|
7QOF
 
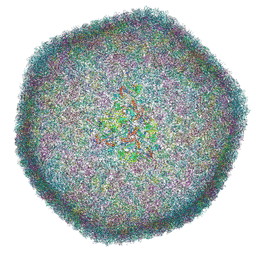 | | Icosahedral capsid of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Head fiber dimer protein gp29, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOG
 
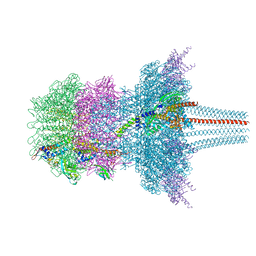 | | Portal protein assembly of the phicrAss001 virion with C12 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, Portal protein gp20, Ring protein 1 gp43, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOH
 
 | | Unique vertex of the phicrAss001 virion with C5 symmetry imposed | | Descriptor: | Auxiliary capsid protein gp36, Head fiber trimer protein gp21, MAGNESIUM ION, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOK
 
 | | Tail muzzle assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | MAGNESIUM ION, Muzzle bound helix, Muzzle protein gp44, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOJ
 
 | | Tail barrel assembly of the phicrAss001 virion with C12 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, MAGNESIUM ION, Portal protein gp20, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOL
 
 | | Tail assembly of the phicrAss001 virion with C6 symmetry imposed | | Descriptor: | Cargo protein 1 gp45, MAGNESIUM ION, Muzzle bound helix, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
7QOI
 
 | | Unique vertex of the phicrAss001 virion | | Descriptor: | Auxiliary capsid protein gp36, Cargo protein 1 gp45, Head fiber trimer protein gp21, ... | | Authors: | Bayfield, O.W, Shkoporov, A.N, Yutin, N, Khokhlova, E.V, Smith, J.L.R, Hawkins, D.E.D.P, Koonin, E.V, Hill, C, Antson, A.A. | | Deposit date: | 2021-12-24 | | Release date: | 2023-03-29 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural atlas of a human gut crassvirus.
Nature, 617, 2023
|
|
1QCO
 
 | | CRYSTAL STRUCTURE OF FUMARYLACETOACETATE HYDROLASE COMPLEXED WITH FUMARATE AND ACETOACETATE | | Descriptor: | ACETOACETIC ACID, CALCIUM ION, FUMARIC ACID, ... | | Authors: | Timm, D.E, Mueller, H.A, Bhanumoorthy, P, Harp, J.M, Bunick, G.J. | | Deposit date: | 1999-05-17 | | Release date: | 2000-06-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of a carbon-carbon bond hydrolase.
Structure Fold.Des., 7, 1999
|
|
4R71
 
 | | Structure of the Qbeta holoenzyme complex in the P1211 crystal form | | Descriptor: | 30S ribosomal protein S1, Elongation factor Ts, Elongation factor Tu, ... | | Authors: | Gytz, H, Seweryn, P, Kutlubaeva, Z, Chetverin, A.B, Brodersen, D.E, Knudsen, C.R. | | Deposit date: | 2014-08-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for RNA-genome recognition during bacteriophage Q beta replication.
Nucleic Acids Res., 43, 2015
|
|
1MT1
 
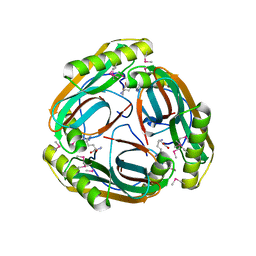 | | The Crystal Structure of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannaschii | | Descriptor: | AGMATINE, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE ALPHA CHAIN, PYRUVOYL-DEPENDENT ARGININE DECARBOXYLASE BETA CHAIN | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-09-20 | | Release date: | 2003-03-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1RO9
 
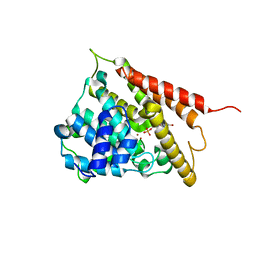 | | CRYSTAL STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHODIESTERASE 4B2B COMPLEXED WITH 8-Br-AMP | | Descriptor: | 8-BROMO-ADENOSINE-5'-MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4B | | Authors: | Xu, R.X, Rocque, W.J, Lambert, M.H, Vanderwall, D.E, Nolte, R.T. | | Deposit date: | 2003-12-01 | | Release date: | 2004-12-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram.
J.Mol.Biol., 337, 2004
|
|
1ROR
 
 | | CRYSTAL STRUCTURES OF THE CATALYTIC DOMAIN OF PHOSPHODIESTERASE 4B2B COMPLEXED WITH AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ZINC ION, cAMP-specific 3',5'-cyclic phosphodiesterase 4B | | Authors: | Xu, R.X, Rocque, W.J, Lambert, M.H, Vanderwall, D.E, Nolte, R.T. | | Deposit date: | 2003-12-02 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram.
J.Mol.Biol., 337, 2004
|
|
1N2M
 
 | | The S53A Proenzyme Structure of Methanococcus jannaschii. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Pyruvoyl-dependent arginine decarboxylase | | Authors: | Tolbert, W.D, Graham, D.E, White, R.H, Ealick, S.E. | | Deposit date: | 2002-10-23 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyruvoyl-Dependent Arginine Decarboxylase from Methanococcus jannaschii:
Crystal Structures of the Self-Cleaved and S53A Proenzyme Forms
Structure, 11, 2003
|
|
1RIQ
 
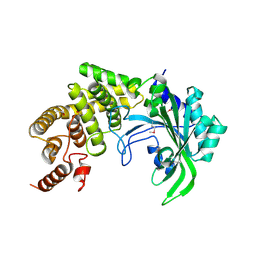 | | The crystal structure of the catalytic fragment of the alanyl-tRNA synthetase | | Descriptor: | Alanyl-tRNA synthetase | | Authors: | Swairjo, M.A, Otero, F.J, Yang, X.-L, Lovato, M.A, Skene, R.J, McRee, D.E, Ribas de Pouplana, L, Schimmel, P. | | Deposit date: | 2003-11-17 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Alanyl-tRNA Synthetase Crystal Structure and Design for Acceptor-Stem Recognition
Mol.Cell, 13, 2004
|
|
7ONG
 
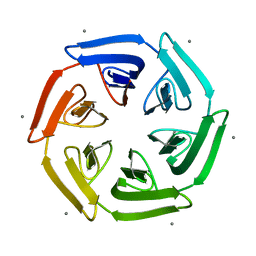 | | Crystal structure of the computationally designed SAKe6BE-L1 protein | | Descriptor: | CALCIUM ION, SAKe6BE-L1 | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ON8
 
 | |
7OPU
 
 | |
7ONA
 
 | | Crystal structure of the computationally designed SAKe6AC protein | | Descriptor: | CALCIUM ION, SAKe6AC | | Authors: | Wouters, S.M.L, Noguchi, H, Velpula, G, Clarke, D.E, Voet, A.R.D, De Feyter, S. | | Deposit date: | 2021-05-25 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAKe: Computationally Designed Modular Protein Building Blocks for Macromolecular Assemblies
To be published
|
|
7ON7
 
 | |
