2XGV
 
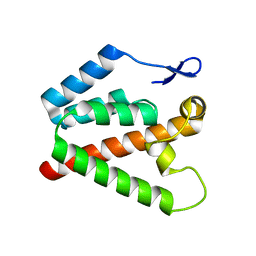 | | Structure of the N-terminal domain of capsid protein from Rabbit Endogenous Lentivirus (RELIK) | | Descriptor: | PSIV CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Robertson, L.E, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2010-06-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
2XQN
 
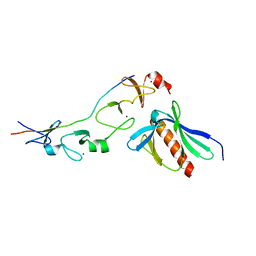 | | Complex of the 2nd and 3rd LIM domains of TES with the EVH1 DOMAIN of MENA and the N-Terminal domain of actin-like protein Arp7A | | Descriptor: | ACTIN-LIKE PROTEIN 7A, ENABLED HOMOLOG, TESTIN, ... | | Authors: | Knowles, P.P, Briggs, D.C, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2010-09-03 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Molecular recognition of the Tes LIM2-3 domains by the actin-related protein Arp7A.
J. Biol. Chem., 286, 2011
|
|
3A0E
 
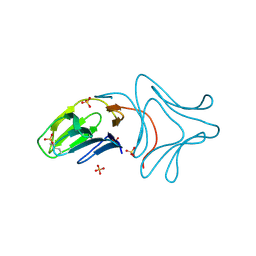 | | Crystal Structure of Polygonatum cyrtonema lectin (PCL) complexed with dimannoside | | Descriptor: | Mannose/sialic acid-binding lectin, SULFATE ION, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2009-03-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of a novel anti-HIV mannose-binding lectin from Polygonatum cyrtonema Hua with unique ligand-binding property and super-structure
J.Struct.Biol., 171, 2010
|
|
3A0D
 
 | |
3NI2
 
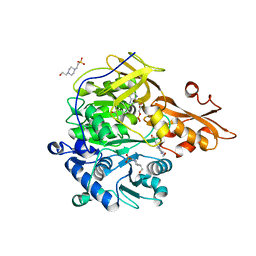 | | Crystal structures and enzymatic mechanisms of a Populus tomentosa 4-coumarate:CoA ligase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-coumarate:CoA ligase, 5'-O-{(S)-hydroxy[3-(4-hydroxyphenyl)propoxy]phosphoryl}adenosine | | Authors: | Hu, Y, Yin, L, Gai, Y, Wang, X.X, Wang, D.C. | | Deposit date: | 2010-06-14 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms
Plant Cell, 22, 2010
|
|
2XQB
 
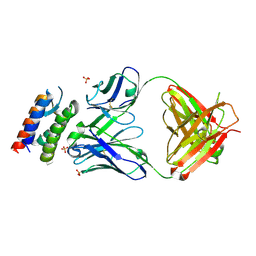 | | Crystal Structure of anti-IL-15 Antibody in Complex with human IL-15 | | Descriptor: | ANTI-IL-15 ANTIBODY, INTERLEUKIN 15, SULFATE ION | | Authors: | Lowe, D.C, Gerhardt, S, Ward, A, Hargreaves, D, Anderson, M, StGallay, S, Vousden, K, Ferraro, F, Pauptit, R.A, Cochrane, D, Pattison, D.V, Buchanan, C, Popovic, B, Finch, D.K, Wilkinson, T, Sleeman, M, Vaughan, T.J, Cruwys, S, Mallinder, P.R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering a High Affinity Anti-Il-15 Antibody: Crystal Structure Reveals an Alpha-Helix in Vh Cdr3 as Key Component of Paratope.
J.Mol.Biol., 406, 2011
|
|
3A0C
 
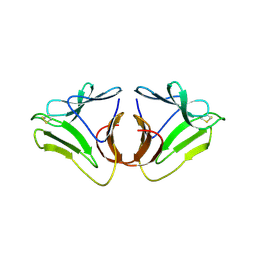 | |
2XPX
 
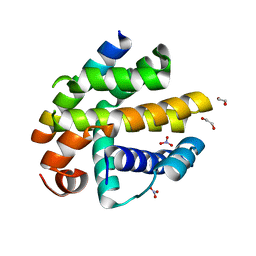 | | Crystal structure of BHRF1:Bak BH3 complex | | Descriptor: | 1,2-ETHANEDIOL, APOPTOSIS REGULATOR BHRF1, BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2010-08-31 | | Release date: | 2011-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis for Apoptosis Inhibition by Epstein-Barr Virus Bhrf1.
Plos Pathog., 6, 2010
|
|
3MIW
 
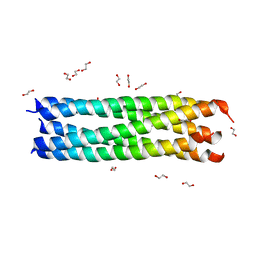 | | Crystal Structure of Rotavirus NSP4 | | Descriptor: | 1,2-ETHANEDIOL, Non-structural glycoprotein 4 | | Authors: | Chacko, A.R, Read, R.J, Dodson, E.J, Rao, D.C, Suguna, K. | | Deposit date: | 2010-04-12 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new pentameric structure of rotavirus NSP4 revealed by molecular replacement.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2XGY
 
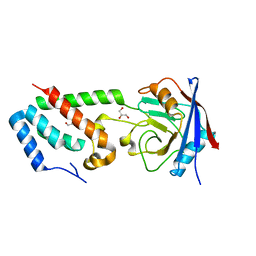 | | Complex of Rabbit Endogenous Lentivirus (RELIK)Capsid with Cyclophilin A | | Descriptor: | GLYCEROL, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A, RELIK CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Robertson, L.E, Haire, L.F, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2010-06-08 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
2XTX
 
 | | Structure of QnrB1 (M102R-Trypsin Treated), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
2WTN
 
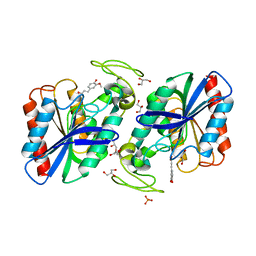 | | Ferulic Acid bound to Est1E from Butyrivibrio proteoclasticus | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, EST1E, GLYCEROL, ... | | Authors: | Goldstone, D.C, Arcus, V.L. | | Deposit date: | 2009-09-17 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Characterization of a Promiscuous Feruloyl Esterase (Est1E) from the Rumen Bacterium Butyrivibrio Proteoclasticus.
Proteins, 78, 2010
|
|
2X1F
 
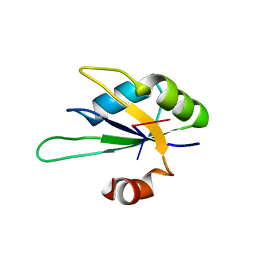 | | Structure of Rna15 RRM with bound RNA (GU) | | Descriptor: | 5'-R(*GP*UP*UP*GP*UP)-3', MRNA 3'-END-PROCESSING PROTEIN RNA15 | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2XFV
 
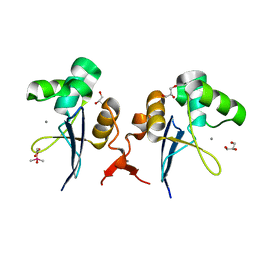 | | Structure of the amino-terminal domain from the cell-cycle regulator Swi6 | | Descriptor: | ACETATE ION, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Smerdon, S.J, Goldstone, D.C, Taylor, I.A. | | Deposit date: | 2010-05-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Amino-Terminal Domain from the Cell-Cycle Regulator Swi6S
Proteins, 78, 2010
|
|
2XH6
 
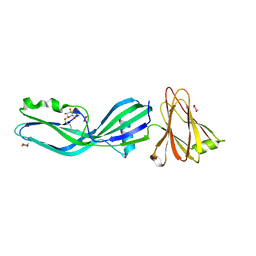 | | Clostridium perfringens enterotoxin | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, HEAT-LABILE ENTEROTOXIN B CHAIN, octyl beta-D-glucopyranoside | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, MCClane, B.A, Basak, A.K. | | Deposit date: | 2010-06-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
2ZGK
 
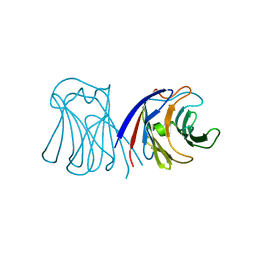 | | Crystal structure of wildtype AAL | | Descriptor: | Anti-tumor lectin | | Authors: | Yang, N, Li, D.F, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZGU
 
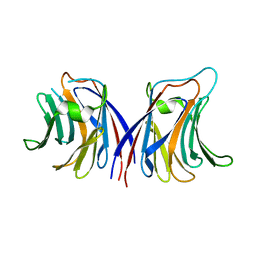 | |
2ZGM
 
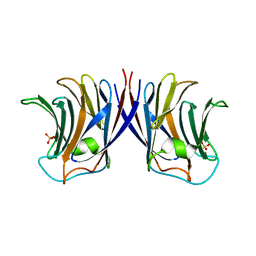 | | Crystal structure of recombinant Agrocybe aegerita lectin,rAAL, complex with lactose | | Descriptor: | Anti-tumor lectin, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Li, D.F, Yang, N, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZGT
 
 | |
2ZGQ
 
 | |
3O6B
 
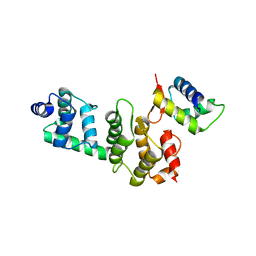 | | A Dual E3 Mechanism for Rub1 Ligation to Cdc53: Dcn1(P)-Cdc53(WHB) low resolution | | Descriptor: | Cell division control protein 53, Defective in cullin neddylation protein 1 | | Authors: | Scott, D.C, Monda, J.K, Grace, C.R.R, Duda, D.M, Kriwacki, R.W, Kurz, T, Schulman, B.A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A dual E3 mechanism for Rub1 ligation to Cdc53.
Mol.Cell, 39, 2010
|
|
3AJM
 
 | | Crystal structure of programmed cell death 10 in complex with inositol 1,3,4,5-tetrakisphosphate | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Programmed cell death protein 10 | | Authors: | Ding, J, Wang, D.C. | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human programmed cell death 10 complexed with inositol-(1,3,4,5)-tetrakisphosphate: a novel adaptor protein involved in human cerebral cavernous malformation.
Biochem.Biophys.Res.Commun., 399, 2010
|
|
3MIN
 
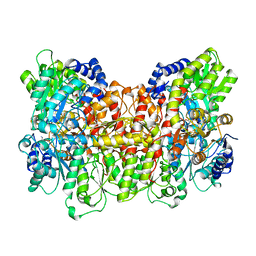 | | NITROGENASE MOFE PROTEIN FROM AZOTOBACTER VINELANDII, OXIDIZED STATE | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Peters, J.W, Stowell, M.H.B, Soltis, S.M, Day, M.W, Kim, J, Rees, D.C. | | Deposit date: | 1996-12-20 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Redox-dependent structural changes in the nitrogenase P-cluster.
Biochemistry, 36, 1997
|
|
3LZF
 
 | |
3AQS
 
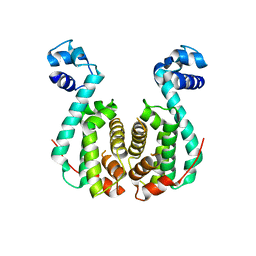 | | Crystal structure of RolR (NCGL1110) without ligand | | Descriptor: | Bacterial regulatory proteins, tetR family | | Authors: | Li, D.F, Zhang, N, Hou, Y.J, Liu, S.J, Wang, D.C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-07-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structures of the transcriptional repressor RolR reveals a novel recognition mechanism between inducer and regulator.
Plos One, 6, 2011
|
|
