4KEF
 
 | |
4KKJ
 
 | | Crystal Structure of Haptocorrin in Complex with Cbi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COB(II)INAMIDE, CYANIDE ION, ... | | Authors: | Furger, E, Frei, D.C, Schibli, R, Fischer, E, Prota, A.E. | | Deposit date: | 2013-05-06 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for universal corrinoid recognition by the cobalamin transport protein haptocorrin.
J.Biol.Chem., 288, 2013
|
|
2X1B
 
 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
4M0V
 
 | | Crystal structure of E.coli SbcD with Mn2+ | | Descriptor: | Exonuclease subunit SbcD, GLYCEROL, MANGANESE (II) ION | | Authors: | Liu, S, Tian, L.F, Yan, X.X, Liang, D.C. | | Deposit date: | 2013-08-02 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4I9U
 
 | | Crystal structure of rabbit LDHA in complex with a fragment inhibitor AP26256 | | Descriptor: | 6-({2-[(5-chloro-2-methoxyphenyl)amino]-2-oxoethyl}sulfanyl)pyridine-3-carboxylic acid, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Kohlmann, A, Stephan, Z.G, Commodore, L, Greenfield, M.T, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
4M6L
 
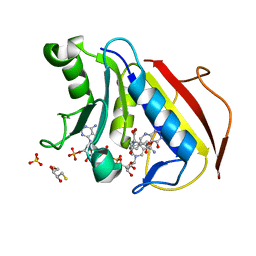 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and 5,10-dideazatetrahydrofolic acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydrofolate reductase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4M6J
 
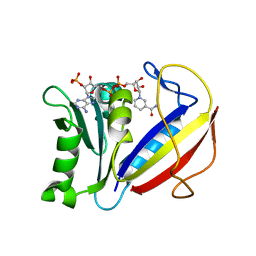 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADPH | | Descriptor: | Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2WH6
 
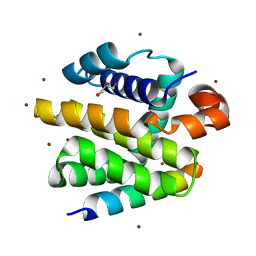 | | Crystal structure of anti-apoptotic BHRF1 in complex with the Bim BH3 domain | | Descriptor: | BCL-2-LIKE PROTEIN 11, BROMIDE ION, EARLY ANTIGEN PROTEIN R, ... | | Authors: | Kvansakul, M, Huang, D.C.S, Colman, P.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1.
PLoS Pathog., 6, 2010
|
|
2WNO
 
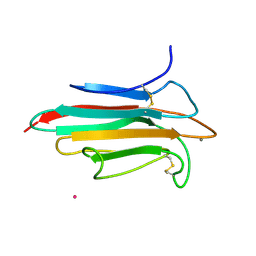 | | X-ray Structure of CUB_C domain from TSG-6 | | Descriptor: | CALCIUM ION, COBALT (II) ION, TUMOR NECROSIS FACTOR-INDUCIBLE GENE 6 PROTEIN | | Authors: | Briggs, D.C, Day, A.J. | | Deposit date: | 2009-07-13 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal Ion-Dependent Heavy Chain Transfer Activity of Tsg-6 Mediates Assembly of the Cumulus-Oocyte Matrix.
J.Biol.Chem., 290, 2015
|
|
4KED
 
 | |
4KQV
 
 | | Topoisomerase iv atp binding domain of francisella tularensis in complex with a small molecule inhibitor | | Descriptor: | 6-fluoro-4-[(3aR,6aR)-hexahydropyrrolo[3,4-b]pyrrol-5(1H)-yl]-N-methyl-2-[(2-methylpyrimidin-5-yl)oxy]-9H-pyrimido[4,5-b]indol-8-amine, IODIDE ION, Topoisomerase IV, ... | | Authors: | Bensen, D.C, Akers-Rodriguez, S, Lam, T, Tari, L.W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A new class of type iia topoisomerase inhibitors with
broad-spectrum antibacterial activity
To be Published
|
|
4M6K
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
2YMY
 
 | | Structure of the murine Nore1-Sarah domain | | Descriptor: | CADMIUM ION, RAS ASSOCIATION DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Makbul, C, Schwarz, D, Aruxandei, D.C, Wolf, E, Hofmann, E, Herrmann, C. | | Deposit date: | 2012-10-10 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Thermodynamic Characterization of Nore1-Sarah: A Small, Helical Module Important in Signal Transduction Networks.
Biochemistry, 52, 2013
|
|
3LFK
 
 | | A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators | | Descriptor: | CITRIC ACID, MarR Like Protein, TVG0766549, ... | | Authors: | Liu, Z, Walton, T.A, Rees, D.C. | | Deposit date: | 2010-01-17 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A reported archaeal mechanosensitive channel is a structural homolog of MarR-like transcriptional regulators.
Protein Sci., 19, 2010
|
|
2Y4Z
 
 | | Structure of the amino-terminal capsid restriction escape mutation N- MLV L10W | | Descriptor: | CAPSID PROTEIN P30, GLYCEROL | | Authors: | Goldstone, D.C, Holden-Dye, K, Ohkura, S, Stoye, J.P, Taylor, I.A. | | Deposit date: | 2011-01-11 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Novel Escape Mutants Suggest an Extensive Trim5Alpha Binding Site Spanning the Entire Outer Surface of the Murine Leukemia Virus Capsid Protein.
Plos Pathog., 7, 2011
|
|
2XGU
 
 | | Structure of the N-terminal domain of capsid protein from Rabbit Endogenous Lentivirus (RELIK) | | Descriptor: | ACETATE ION, RELIK CAPSID N-TERMINAL DOMAIN | | Authors: | Goldstone, D.C, Taylor, I.A, Robertson, L.E, Haire, L.F, Stoye, J.P. | | Deposit date: | 2010-06-07 | | Release date: | 2010-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural and Functional Analysis of Prehistoric Lentiviruses Uncovers an Ancient Molecular Interface.
Cell Host Microbe, 8, 2010
|
|
2ZGN
 
 | | crystal structure of recombinant Agrocybe aegerita lectin, rAAL, complex with galactose | | Descriptor: | Anti-tumor lectin, beta-D-galactopyranose | | Authors: | Yang, N, Li, D.F, Wang, D.C. | | Deposit date: | 2008-01-23 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the tumor cell apoptosis-inducing activity of an antitumor lectin from the edible mushroom Agrocybe aegerita
J.Mol.Biol., 387, 2009
|
|
2ZJT
 
 | |
3LYZ
 
 | |
2ZGS
 
 | |
2ZGR
 
 | |
2XTY
 
 | | Structure of QnrB1 (R167E-Trypsin Treated), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1 | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-13 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
2XTW
 
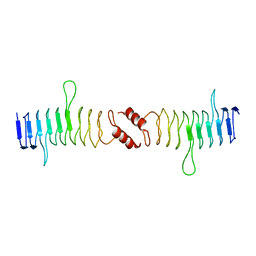 | | Structure of QnrB1 (Full length), a plasmid-mediated fluoroquinolone resistance protein | | Descriptor: | QNRB1 | | Authors: | Vetting, M.W, Hegde, S.S, Park, C.H, Jacoby, G.A, Hooper, D.C, Blanchard, J.S. | | Deposit date: | 2010-10-12 | | Release date: | 2010-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of Qnrb1, a Plasmid-Mediated Fluoroquinolone Resistance Factor.
J.Biol.Chem., 286, 2011
|
|
2YHJ
 
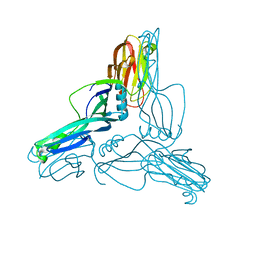 | | Clostridium perfringens Enterotoxin at 4.0 Angstrom Resolution | | Descriptor: | HEAT-LABILE ENTEROTOXIN B CHAIN | | Authors: | Briggs, D.C, Naylor, C.E, Smedley III, J.G, McClane, B.A, Basak, A.K. | | Deposit date: | 2011-05-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of the Food-Poisoning Clostridium Perfringens Enterotoxin Reveals Similarity to the Aerolysin-Like Pore-Forming Toxins
J.Mol.Biol., 413, 2011
|
|
2Y6T
 
 | | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia pestis | | Descriptor: | CHYMOTRYPSINOGEN A, ECOTIN, SULFATE ION | | Authors: | Clark, E.A, Walker, N, Ford, D.C, Cooper, I.A, Oyston, P.C.F, Acharya, K.R. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia Pestis.
J.Biol.Chem., 286, 2011
|
|
