1OG5
 
 | | Structure of human cytochrome P450 CYP2C9 | | Descriptor: | CYTOCHROME P450 2C9, HEME C, S-WARFARIN | | Authors: | Williams, P.A, Cosme, J, Ward, A, Angove, H.C, Matak Vinkovic, D, Jhoti, H. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Human Cytochrome P450 2C9 with Bound Warfarin
Nature, 424, 2003
|
|
2YOF
 
 | | Plasmodium falciparum thymidylate kinase in complex with a (thio)urea- beta-deoxythymidine inhibitor | | Descriptor: | 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-[[(2R,3S)-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-oxidanyl-oxolan-2-yl]methyl]thiourea, ACETATE ION, THYMIDYLATE KINASE, ... | | Authors: | Huaqing, C, Carrero-Lerida, J, Silva, A.P.G, Whittingham, J.L, Brannigan, J.A, Ruiz-Perez, L.M, Read, K.D, Wilson, K.S, Gonzalez-Pacanowska, D, Gilbert, I.H. | | Deposit date: | 2012-10-24 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Synthesis and Evaluation of Alpha-Thymidine Analogues as Novel Antimalarials.
J.Med.Chem., 55, 2012
|
|
2YE0
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise (K206A mutant) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Goedhart, J, Noirclerc-Savoye, M, Lelimousin, M, Joosen, L, Hink, M.A, van Weeren, L, Gadella, T.W.J, Royant, A. | | Deposit date: | 2011-03-25 | | Release date: | 2012-03-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structure-Guided Evolution of Cyan Fluorescent Proteins Towards a Quantum Yield of 93%
Nat.Commun, 3, 2012
|
|
1OPJ
 
 | | Structural basis for the auto-inhibition of c-Abl tyrosine kinase | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, MYRISTIC ACID, ... | | Authors: | Nagar, B, Hantschel, O, Young, M.A, Scheffzek, K, Veach, D, Bornmann, W, Clarkson, B, Superti-Furga, G, Kuriyan, J. | | Deposit date: | 2003-03-06 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the autoinhibition of c-Abl tyrosine kinase
Cell(Cambridge,Mass.), 112, 2003
|
|
1OIG
 
 | | The solution structure of the DPY module from the Dumpy protein | | Descriptor: | Dumpy, isoform Y | | Authors: | Wilkin, M.B, Becker, M.N, Mulvey, D, Phan, I, Chao, A, Cooper, K, Chung, H.J, Campbell, I.D, Baron, M, MacIntyre, R. | | Deposit date: | 2003-06-18 | | Release date: | 2003-06-26 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Drosophila Dumpy is a Gigantic Extracellular Protein Required to Maintain Tension at Epidermal-Cuticle Attachment Sites
Curr.Biol., 10, 2000
|
|
1OHQ
 
 | | Crystal structure of HEL4, a soluble human VH antibody domain resistant to aggregation | | Descriptor: | IMMUNOGLOBULIN | | Authors: | Jespers, L, Schon, O, James, L.C, Veprintsev, D, Winter, G. | | Deposit date: | 2003-05-30 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hel4, a Soluble, Refoldable Human V(H) Single Domain with a Germ-Line Scaffold
J.Mol.Biol., 337, 2004
|
|
1OM2
 
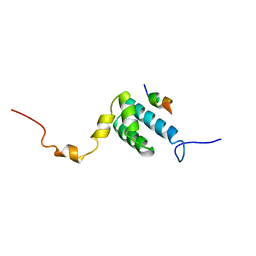 | | SOLUTION NMR STRUCTURE OF THE MITOCHONDRIAL PROTEIN IMPORT RECEPTOR TOM20 FROM RAT IN A COMPLEX WITH A PRESEQUENCE PEPTIDE DERIVED FROM RAT ALDEHYDE DEHYDROGENASE (ALDH) | | Descriptor: | PROTEIN (MITOCHONDRIAL ALDEHYDE DEHYDROGENASE), PROTEIN (MITOCHONDRIAL IMPORT RECEPTOR SUBUNIT TOM20) | | Authors: | Abe, Y, Shodai, T, Muto, T, Mihara, K, Torii, H, Nishikawa, S, Endo, T, Kohda, D. | | Deposit date: | 1999-04-23 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20.
Cell(Cambridge,Mass.), 100, 2000
|
|
1ONA
 
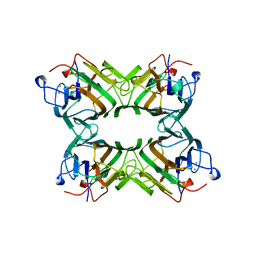 | | CO-CRYSTALS OF CONCANAVALIN A WITH METHYL-3,6-DI-O-(ALPHA-D-MANNOPYRANOSYL)-ALPHA-D-MANNOPYRANOSIDE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION, ... | | Authors: | Bouckaert, J, Maes, D, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1996-07-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structure of the complex between concanavalin A and methyl-3,6-di-O-(alpha-D-mannopyranosyl)-alpha-D-mannopyranoside reveals two binding modes.
J.Biol.Chem., 271, 1996
|
|
1OOE
 
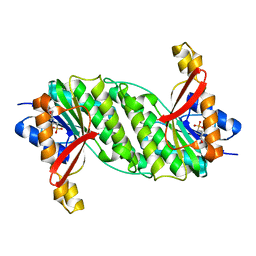 | | Structural Genomics of Caenorhabditis elegans : Dihydropteridine reductase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Dihydropteridine reductase | | Authors: | Symersky, J, Li, S, Nagy, L, Qiu, S, Lin, G, Tsao, J, Luo, D, Carson, M, DeLucas, L, Luo, M, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2003-03-03 | | Release date: | 2003-03-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: structure of dihydropteridine reductase.
Proteins, 53, 2003
|
|
1OP8
 
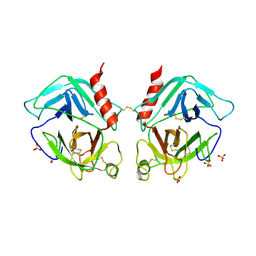 | | Crystal Structure of Human Granzyme A | | Descriptor: | Granzyme A, SULFATE ION | | Authors: | Hink-Schauer, C, Estebanez-Perpina, E, Bode, W, Jenne, D. | | Deposit date: | 2003-03-05 | | Release date: | 2003-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the apoptosis-inducing human granzyme A dimer
NAT.STRUCT.BIOL., 10, 2003
|
|
1OS1
 
 | | Structure of Phosphoenolpyruvate Carboxykinase complexed with ATP,Mg, Ca and pyruvate. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Sudom, A, Walters, R, Pastushok, L, Goldie, D, Prasad, L, Delbaere, L.T, Goldie, H. | | Deposit date: | 2003-03-18 | | Release date: | 2003-09-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanisms of activation of phosphoenolpyruvate carboxykinase from Escherichia coli by Ca2+ and of desensitization by trypsin.
J.BACTERIOL., 185, 2003
|
|
1OUS
 
 | | Lecb (PA-LII) calcium-free | | Descriptor: | SULFATE ION, hypothetical protein LecB | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-25 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1OVS
 
 | | LecB (PA-LII) in complex with core trimannoside | | Descriptor: | CALCIUM ION, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose, ... | | Authors: | Loris, R, Tielker, D, Jaeger, K.-E, Wyns, L. | | Deposit date: | 2003-03-27 | | Release date: | 2003-09-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Carbohydrate Recognition by the Lectin LecB from Pseudomonas aeruginosa
J.MOL.BIOL., 331, 2003
|
|
1P6Y
 
 | | T4 LYSOZYME CORE REPACKING MUTANT M120Y/TA | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-04-30 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
1OOA
 
 | | CRYSTAL STRUCTURE OF NF-kB(p50)2 COMPLEXED TO A HIGH-AFFINITY RNA APTAMER | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit, RNA aptamer | | Authors: | Huang, D.B, Vu, D, Cassiday, L.A, Zimmerman, J.M, Maher III, L.J, Ghosh, G. | | Deposit date: | 2003-03-03 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Q6N
 
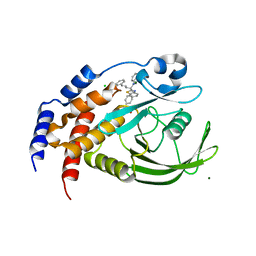 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 4 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1QGB
 
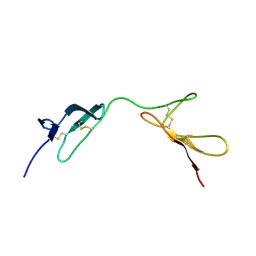 | | SOLUTION STRUCTURE OF THE N-TERMINAL F1 MODULE PAIR FROM HUMAN FIBRONECTIN | | Descriptor: | PROTEIN (FIBRONECTIN) | | Authors: | Potts, J.R, Bright, J.R, Bolton, D, Pickford, A.R, Campbell, I.D. | | Deposit date: | 1999-04-21 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal F1 module pair from human fibronectin.
Biochemistry, 38, 1999
|
|
1QXC
 
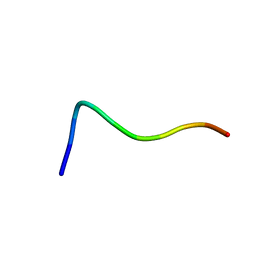 | | NMR structure of the fragment 25-35 of beta amyloid peptide in 20/80 v:v hexafluoroisopropanol/water mixture | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-05 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QI1
 
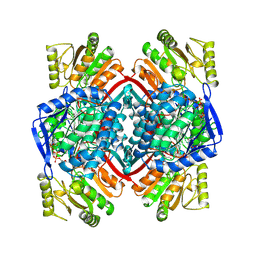 | | Ternary Complex of an NADP Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (NADP-DEPENDENT NONPHOSPHORYLATING GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE), ... | | Authors: | Cobessi, D, Tete-Favier, F, Marchal, S, Branlant, G, Aubry, A. | | Deposit date: | 1999-06-02 | | Release date: | 2001-01-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical investigations of the catalytic mechanism of an NADP-dependent aldehyde dehydrogenase from Streptococcus mutans.
J.Mol.Biol., 300, 2000
|
|
1QWD
 
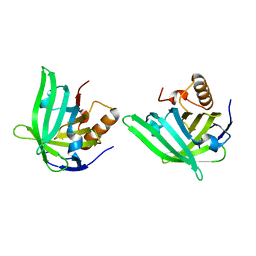 | | CRYSTAL STRUCTURE OF A BACTERIAL LIPOCALIN, THE BLC GENE PRODUCT FROM E. COLI | | Descriptor: | Outer membrane lipoprotein blc | | Authors: | Campanacci, V, Nurizzo, D, Spinelli, S, Valencia, C, Cambillau, C. | | Deposit date: | 2003-09-02 | | Release date: | 2004-04-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of the Escherichia coli lipocalin Blc suggests a possible role in phospholipid binding
Febs Lett., 562, 2004
|
|
1QX4
 
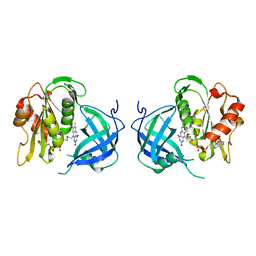 | | Structrue of S127P mutant of cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bewley, M.C, Davis, C.A, Marohnic, C.C, Taormina, D, Barber, M.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the S127P mutant of cytochrome b5 reductase that causes methemoglobinemia shows the AMP moiety of the flavin occupying the substrate binding site
Biochemistry, 42, 2003
|
|
1QUN
 
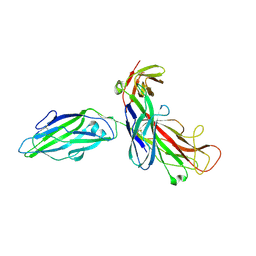 | | X-RAY STRUCTURE OF THE FIMC-FIMH CHAPERONE ADHESIN COMPLEX FROM UROPATHOGENIC E.COLI | | Descriptor: | MANNOSE-SPECIFIC ADHESIN FIMH, PAPD-LIKE CHAPERONE FIMC | | Authors: | Choudhury, D, Thompson, A, Stojanoff, V, Langerman, S, Pinkner, J, Hultgren, S.J, Knight, S. | | Deposit date: | 1999-07-01 | | Release date: | 1999-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli.
Science, 285, 1999
|
|
1QWP
 
 | | NMR analysis of 25-35 fragment of beta amyloid peptide | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-03 | | Release date: | 2004-09-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
1QX8
 
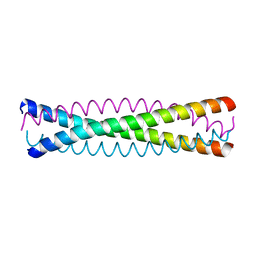 | | Crystal structure of a five-residue deletion mutant of the Rop protein | | Descriptor: | Regulatory protein ROP | | Authors: | Glykos, N.M, Vlassi, M, Papanikolaou, Y, Kotsifaki, D, Cesareni, G, Kokkinidis, M. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Loopless Rop: structure and dynamics of an engineered homotetrameric variant of the repressor of primer protein.
Biochemistry, 45, 2006
|
|
1QYT
 
 | | Solution structure of fragment (25-35) of beta amyloid peptide in SDS micellar solution | | Descriptor: | 11-mer peptide from Amyloid beta A4 protein | | Authors: | D'Ursi, A.M, Armenante, M.R, Guerrini, R, Salvadori, S, Sorrentino, G, Picone, D. | | Deposit date: | 2003-09-12 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of amyloid beta-peptide (25-35) in different media
J.Med.Chem., 47, 2004
|
|
