2J95
 
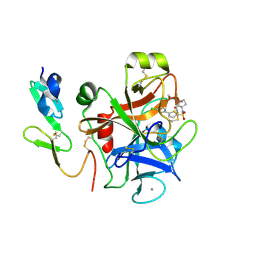 | | CRYSTAL STRUCTURE OF A HUMAN FACTOR XA INHIBITOR COMPLEX | | Descriptor: | 5'-CHLORO-N-{(3S)-1-[(1S)-1-METHYL-2-MORPHOLIN-4-YL-2-OXOETHYL]-2-OXOPYRROLIDIN-3-YL}-2,2'-BITHIOPHENE-5-SULFONAMIDE, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Chan, C, Borthwick, A.D, Brown, D, Campbell, M, Chaudry, L, Chung, C.W, Convery, M.A, Hamblin, J.N, Johnstone, L, Kelly, H.A, Kleanthous, S, Burns-Kurtis, C.L, Patikis, A, Patel, C, Pateman, A.J, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Whitworth, C, Young, R.J, Zhou, P. | | Deposit date: | 2006-11-02 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Factor Xa Inhibitors: S1 Binding Interactions of a Series of N-{(3S)-1-[(1S)-1-Methyl-2-Morpholin-4-Yl-2-Oxoethyl]-2-Oxopyrrolidin-3-Yl}Sulfonamides.
J.Med.Chem., 50, 2007
|
|
3ZYH
 
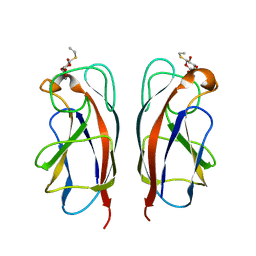 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYB
 
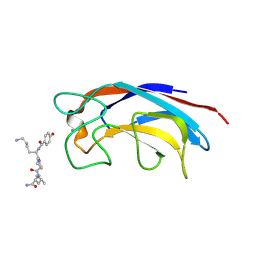 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A RESOLUTION | | Descriptor: | CALCIUM ION, GALA-LYS-PRO-LEUNH2, P-HYDROXYBENZOIC ACID, ... | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZKE
 
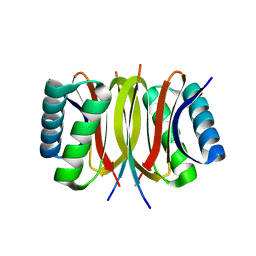 | | Structure of LC8 in complex with Nek9 peptide | | Descriptor: | DYNEIN LIGHT CHAIN 1, CYTOPLASMIC, NEK9 PROTEIN | | Authors: | Gallego, P, Velazquez-Campoy, A, Regue, L, Roig, J, Reverter, D. | | Deposit date: | 2013-01-22 | | Release date: | 2013-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of the Regulation of the Dynll/Lc8 Binding to Nek9 by Phosphorylation
J.Biol.Chem., 288, 2013
|
|
3ZYF
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH NPG AT 1.9 A RESOLUTION | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, CALCIUM ION, PA-I GALACTOPHILIC LECTIN | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
8AS6
 
 | | Structure of the SFTSV L protein bound to 5' cRNA hook [5' HOOK] | | Descriptor: | RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*C)-3'), RNA-dependent RNA-polymerase L protein | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
8AS7
 
 | | Structure of the SFTSV L protein stalled at early elongation [EARLY-ELONGATION] | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, MAGNESIUM ION, ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Milewski, M, Busch, C, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2022-08-18 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into viral genome replication by the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 51, 2023
|
|
5T8N
 
 | |
8AJX
 
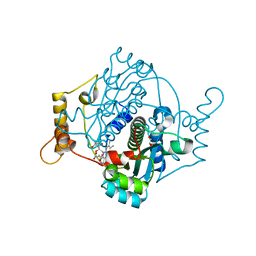 | | E. coli NfsA with Fumarate | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, FUMARIC ACID, ... | | Authors: | Day, M.A, Jarrom, D, White, S.A, Hyde, E.I. | | Deposit date: | 2022-07-28 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Oxygen-insensitive nitroreductase E. coli NfsA, but not NfsB, is inhibited by fumarate.
Proteins, 91, 2023
|
|
3ZRA
 
 | | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators | | Descriptor: | N-{(1R)-1-[4-(2-CHLORO-5-FLUOROPYRIDIN-3-YL)PHENYL]ETHYL}-3,5-DIMETHYLISOXAZOLE-4-SULFONAMIDE, PROGESTERONE RECEPTOR, SULFATE ION | | Authors: | Lusher, S.J, Raaijmakers, H.C.A, Vu-Pham, D, Dechering, K, Wai Lam, T, Brown, A.R, Hamilton, N.M, Nimz, O, Azevedo, R, McGuire, R, Oubrie, A, de Vlieg, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for agonism and antagonism for a set of chemically related progesterone receptor modulators.
J. Biol. Chem., 286, 2011
|
|
3DLJ
 
 | | Crystal structure of human carnosine dipeptidase 1 | | Descriptor: | Beta-Ala-His dipeptidase, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dobrovetsky, E, Seitova, A, He, H, Tempel, W, Kozieradzki, I, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Bochkarev, A, Cossar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-06-27 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of human carnosine dipeptidase 1.
To be Published
|
|
7KVC
 
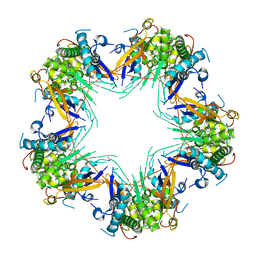 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (decamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
7KVD
 
 | | Cryo-EM structure of Mal de Rio Cuarto virus P9-1 viroplasm protein (dodecamer) | | Descriptor: | p9-1 | | Authors: | Llauger, G, Melero, R, Monti, D, Sycz, G, Huck-Iriart, C, Cerutti, M.L, Klinke, S, Arranz, R, Carazo, J.M, Goldbaum, F.A, del Vas, M, Otero, L.H. | | Deposit date: | 2020-11-27 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | A Fijivirus Major Viroplasm Protein Shows RNA-Stimulated ATPase Activity by Adopting Pentameric and Hexameric Assemblies of Dimers.
Mbio, 14, 2023
|
|
8AI7
 
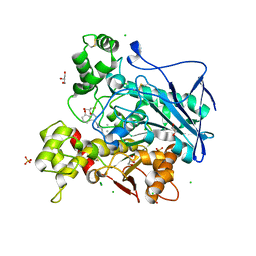 | | Structure of carbamoylated human butyrylcholinesterase upon reaction with 3-(((2-cycloheptylethyl)(methyl)amino)methyl)-1H-indol-7-yl N,N-dimethylcarbamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[[2-cycloheptylethyl(methyl)amino]methyl]-1~{H}-indol-7-ol, ... | | Authors: | Brazzolotto, X, Meden, A, Knez, D, Gobec, S, Nachon, F. | | Deposit date: | 2022-07-25 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Pseudo-irreversible butyrylcholinesterase inhibitors: Structure-activity relationships, computational and crystallographic study of the N-dialkyl O-arylcarbamate warhead.
Eur.J.Med.Chem., 247, 2023
|
|
3ZV0
 
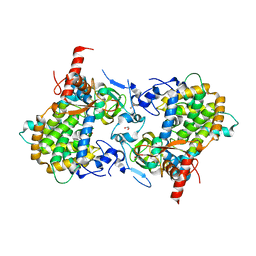 | | Structure of the SHQ1P-CBF5P complex | | Descriptor: | GLYCEROL, H/ACA RIBONUCLEOPROTEIN COMPLEX SUBUNIT 4, PROTEIN SHQ1 | | Authors: | Walbott, H, Machado-Pinilla, R, Liger, D, Blaud, M, Rety, S, Grozdanov, P.N, Godin, K, vanTilbeurgh, H, Varani, G, Meier, U.T, Leulliot, N. | | Deposit date: | 2011-07-22 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The H/Aca Rnp Assembly Factor Shq1 Functions as an RNA Mimic.
Genes Dev., 25, 2011
|
|
5TBF
 
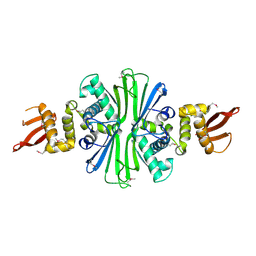 | |
2IUP
 
 | |
3ZWL
 
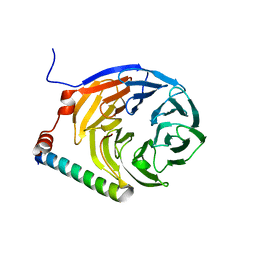 | |
8AJ7
 
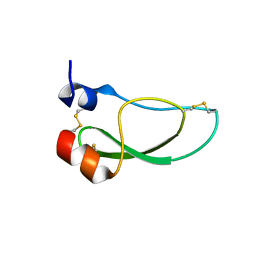 | | Kunitz domain of Amblyomin-X | | Descriptor: | 1,2-ETHANEDIOL, Kunitz domain of Amblyomin-X | | Authors: | Ciccone, L, Servent, D, Stura, E.A. | | Deposit date: | 2022-07-27 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional properties of the Kunitz-type and C-terminal domains of Amblyomin-X supporting its antitumor activity.
Front Mol Biosci, 10, 2023
|
|
3ZXX
 
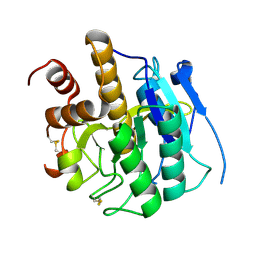 | | Structure of self-cleaved protease domain of PatA | | Descriptor: | SUBTILISIN-LIKE PROTEIN | | Authors: | Koehnke, J, Zollman, D, Vendome, J, Raab, A, Houssen, W.E, Smith, M.C, Jaspars, M, Naismith, J.H. | | Deposit date: | 2011-08-16 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Discovery of New Cyanobactins from Cyanothece Pcc 7425 Defines a New Signature for Processing of Patellamides.
Chembiochem, 13, 2012
|
|
8B0F
 
 | | CryoEM structure of C5b8-CD59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bubeck, D, Couves, E.C, Gardner, S. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
8B0G
 
 | | 2C9, C5b9-CD59 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD59 glycoprotein, Complement C5, ... | | Authors: | Couves, E.C, Gardner, S, Bubeck, D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for membrane attack complex inhibition by CD59.
Nat Commun, 14, 2023
|
|
5TIF
 
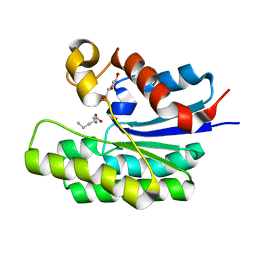 | | x-ray structure of acyl-CoA thioesterase I, TesA, triple mutant M141L/Y145K/L146K in complex with octanoic acid | | Descriptor: | Acyl-CoA thioesterase I, DI(HYDROXYETHYL)ETHER, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Thoden, J.B, Holden, H.M, Grisewood, M.J, Hernandez Lozada, N.J, Gifford, N.P, Mendez-Perez, D, Schoenberger, H.A, Allan, M.F, Pfleger, B.F, Marines, C.D. | | Deposit date: | 2016-10-02 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Computational Redesign of Acyl-ACP Thioesterase with Improved Selectivity toward Medium-Chain-Length Fatty Acids.
ACS Catal, 7, 2017
|
|
8A9B
 
 | |
5T4Q
 
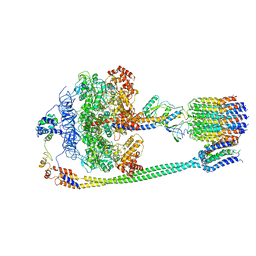 | | Autoinhibited E. coli ATP synthase state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Smits, C, Wong, A.S.W, Ishmukhametov, R, Stock, D, Sandin, S, Stewart, A.G. | | Deposit date: | 2016-08-29 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (8.53 Å) | | Cite: | Cryo-EM structures of the autoinhibitedE. coliATP synthase in three rotational states.
Elife, 5, 2016
|
|
