6WU2
 
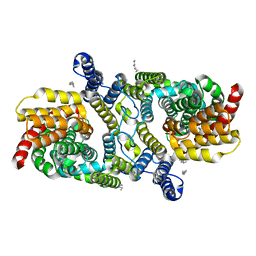 | | Structure of the LaINDY-malate complex | | Descriptor: | DASS family sodium-coupled anion symporter, DECANE, HEXANE, ... | | Authors: | Sauer, D.B, Marden, J.J, Cocco, N, Song, J.M, Wang, D.N, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis for the reaction cycle of DASS dicarboxylate transporters.
Elife, 9, 2020
|
|
8QA2
 
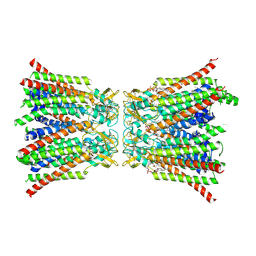 | |
8QA3
 
 | |
6FVL
 
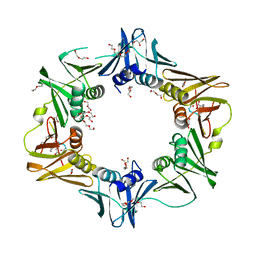 | | DNA polymerase sliding clamp from Escherichia coli with bound P7 peptide | | Descriptor: | Beta sliding clamp, GLYCEROL, P7 peptide, ... | | Authors: | Martiel, I, Andre, C, Olieric, V, Guichard, G, Burnouf, D. | | Deposit date: | 2018-03-04 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | Peptide Interactions on Bacterial Sliding Clamps.
Acs Infect Dis., 2019
|
|
5AQP
 
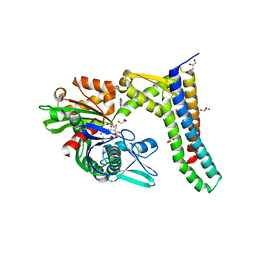 | | Fragment-based screening of HSP70 sheds light on the functional role of ATP-binding site residues | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BAG FAMILY MOLECULAR CHAPERONE REGULATOR 1, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, A.M, Westwood, I.M, Osborne, J.D, Matthews, T.P, Cheeseman, M.D, Rowlands, M.G, Jeganathan, F, Burke, R, Lee, D, Kadi, N, Liu, M, Richards, M, McAndrew, C, Yahya, N, Dobson, S.E, Jones, K, Workman, P, Collins, I, van Montfort, R.L.M. | | Deposit date: | 2015-09-22 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fragment-based approach applied to a highly flexible target: Insights and challenges towards the inhibition of HSP70 isoforms.
Sci Rep, 6, 2016
|
|
8QA0
 
 | |
8UXN
 
 | | Caulobacter crescentus FljM flagellar filament (symmetrized) | | Descriptor: | Flagellin FljM | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2023-11-09 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
6WCJ
 
 | | Asymmetric vertex of the clathrin minicoat cage | | Descriptor: | Clathrin heavy chain 1, Clathrin light chain B | | Authors: | Paraan, M, Mendez, J, Sharum, S, Kurtin, D, He, H, Stagg, S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | The structures of natively assembled clathrin-coated vesicles.
Sci Adv, 6, 2020
|
|
8V47
 
 | |
8UXJ
 
 | | Caulobacter crescentus FljK flagellar filament (asymmetrical) | | Descriptor: | Flagellin FljK | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2023-11-09 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Atomic-level architecture of Caulobacter crescentus flagellar filaments provide evidence for multi-flagellin filament stabilization
To Be Published
|
|
8V45
 
 | |
6FZW
 
 | |
5TE3
 
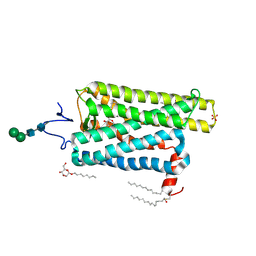 | | Crystal structure of Bos taurus opsin at 2.7 Angstrom | | Descriptor: | PALMITIC ACID, Rhodopsin, SULFATE ION, ... | | Authors: | Gulati, S, Kiser, P.D, Palczewski, K. | | Deposit date: | 2016-09-20 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Photocyclic behavior of rhodopsin induced by an atypical isomerization mechanism.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8QAI
 
 | |
8QAH
 
 | |
8QKD
 
 | |
5NP0
 
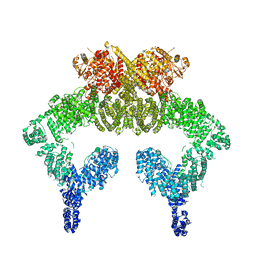 | | Closed dimer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
8V48
 
 | |
6Z3Z
 
 | |
8QAE
 
 | |
6C69
 
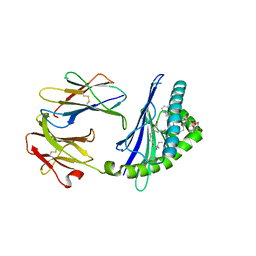 | | Structure of glycolipid aGSA[12,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
6C6E
 
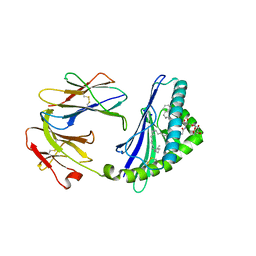 | | Structure of glycolipid aGSA[26,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
5B5K
 
 | | Crystal structure of Izumo1, the mammalian sperm ligand for egg Juno | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Izumo sperm-egg fusion protein 1 | | Authors: | Nishimura, K, Han, L, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-05-11 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of sperm Izumo1 reveals unexpected similarities with Plasmodium invasion proteins.
Curr.Biol., 26, 2016
|
|
8QAG
 
 | |
8V49
 
 | |
