6PCH
 
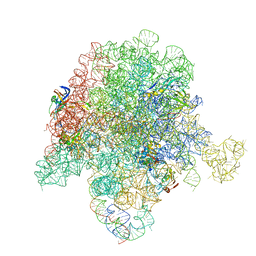 | | E. coli 50S ribosome bound to compound 21 | | Descriptor: | (3R,4R,5E,10E,12E,14S,26aR)-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-8,9,14,15,24,25,26,26a-octahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,16,22(4H,17H)-tetrone, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
8PUZ
 
 | |
4V4Y
 
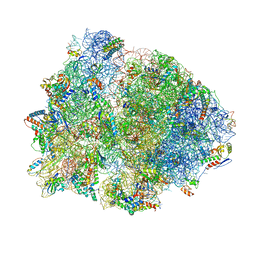 | | Crystal structure of the 70S Thermus thermophilus ribosome with translocated and rotated Shine-Dalgarno Duplex. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Yusupova, G, Rees, B, Moras, D, Yusupov, M. | | Deposit date: | 2006-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural basis for messenger RNA movement on the ribosome.
Nature, 444, 2006
|
|
8PRD
 
 | |
1TGX
 
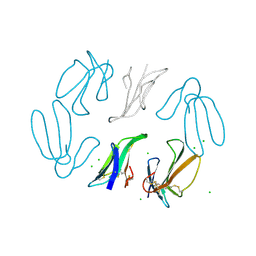 | | X-RAY STRUCTURE AT 1.55 A OF TOXIN GAMMA, A CARDIOTOXIN FROM NAJA NIGRICOLLIS VENOM. CRYSTAL PACKING REVEALS A MODEL FOR INSERTION INTO MEMBRANES | | Descriptor: | CHLORIDE ION, GAMMA-CARDIOTOXIN | | Authors: | Bilwes, A, Rees, B, Moras, D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-04-30 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure at 1.55 A of toxin gamma, a cardiotoxin from Naja nigricollis venom. Crystal packing reveals a model for insertion into membranes.
J.Mol.Biol., 239, 1994
|
|
8PRO
 
 | |
1JXN
 
 | | Crystal Structure of the Lectin I from Ulex europaeus in complex with the methyl glycoside of alpha-L-fucose | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Audette, G.F, Olson, D.J.H, Ross, A.R.S, Quail, J.W, Delbaere, L.T.J. | | Deposit date: | 2001-09-07 | | Release date: | 2002-12-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Examination of the Structural Basis for O(H) Blood Group Specificity by Ulex europaeus Lectin I
Can.J.Chem., 80, 2002
|
|
6VRM
 
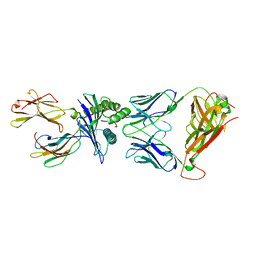 | | T cell receptor-p53-HLA-A2 complex | | Descriptor: | Beta-2-microglobulin, Cellular tumor antigen p53 peptide, MHC class I antigen, ... | | Authors: | Wu, D, Gallagher, D.T, Gowthaman, R, Pierce, B.G, Mariuzza, R.A. | | Deposit date: | 2020-02-08 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for oligoclonal T cell recognition of a shared p53 cancer neoantigen.
Nat Commun, 11, 2020
|
|
2I5G
 
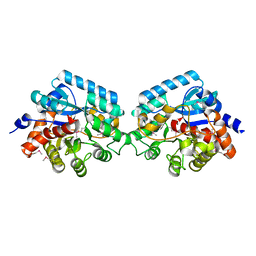 | | Crystal strcuture of amidohydrolase from Pseudomonas aeruginosa | | Descriptor: | amidohydrolase | | Authors: | Min, T, Sauder, J.M, Wasserman, S.R, Smith, D, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-24 | | Release date: | 2006-09-05 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of amidohydrolase from Pseudomonas aeruginosa
To be Published
|
|
5DDO
 
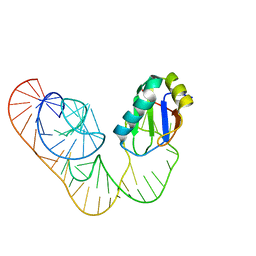 | |
1TLL
 
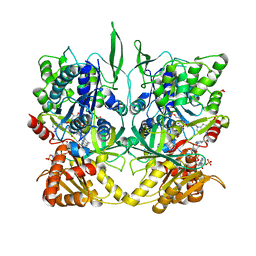 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE REDUCTASE MODULE AT 2.3 A RESOLUTION. | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Garcin, E.D, Bruns, C.M, Lloyd, S.J, Hosfield, D.J, Tiso, M, Gachhui, R, Stuehr, D.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for isozyme-specific regulation of electron transfer in nitric-oxide synthase
J.Biol.Chem., 279, 2004
|
|
7B03
 
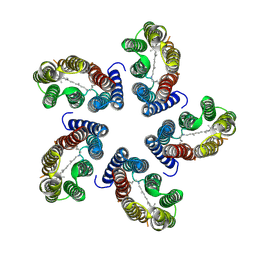 | |
2HY5
 
 | | Crystal structure of DsrEFH | | Descriptor: | DsrH, Intracellular sulfur oxidation protein dsrF, Putative sulfurtransferase dsrE | | Authors: | Shin, D.H, Schulte, A, Dahl, C, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2006-08-04 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of DsrEFH
To be Published
|
|
8TJH
 
 | |
6W47
 
 | | Peptoid-Containing Collagen Peptide | | Descriptor: | 1,2-ETHANEDIOL, Collagen-like peptide | | Authors: | Chenoweth, D.M, Melton, S.D. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Rules for the design of aza-glycine stabilized triple-helical collagen peptides.
Chem Sci, 11, 2020
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5JZK
 
 | | The Structure of Ultra Stable Green Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yong, K.J, Gunn, N.J, Scott, D.J, Griffin, M.D.W. | | Deposit date: | 2016-05-17 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY.
Sci Rep, 8, 2018
|
|
7SOB
 
 | |
4V0G
 
 | | JAK3 in complex with a covalent EGFR inhibitor | | Descriptor: | N-[3-(2-{3-amino-6-[1-(1-methylpiperidin-4-yl)-1H-pyrazol-4-yl]pyrazin-2-yl}-1H-benzimidazol-1-yl)phenyl]propanamide, TYROSINE-PROTEIN KINASE JAK3 | | Authors: | Debreczeni, J.E, Hennessy, E.J, Chuaquini, C, Ashton, S, Coclough, N, Cross, D.A.E, Eberlein, C, Gingipalli, L, Klinowska, T.C.M, Orme, J.P, Sha, L, Wu, X. | | Deposit date: | 2014-09-16 | | Release date: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Utilisation of Structure Based Design to Identify Novel, Irreversible Inhibitors of the Epidermal Growth Factor Receptor (Egfr) Harboring the Gatekeeper T790M Mutation
To be Published
|
|
2I9U
 
 | |
5K54
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
7ZPJ
 
 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 isoform 1 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
8VM8
 
 | |
8VMA
 
 | |
8VM9
 
 | |
