2W1B
 
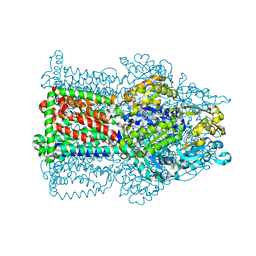 | | The structure of the efflux pump AcrB in complex with bile acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, ACRIFLAVIN RESISTANCE PROTEIN B | | Authors: | Drew, D, Klepsch, M.M, Newstead, S, Flaig, R, De Gier, J.W, Iwata, S, Beis, K. | | Deposit date: | 2008-10-17 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | The Structure of the Efflux Pump Acrb in Complex with Bile Acid.
Mol.Membr.Biol., 25, 2008
|
|
2OUY
 
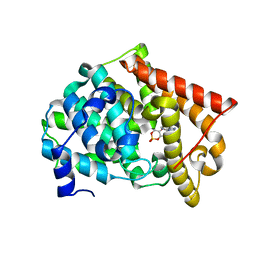 | | crystal structure of pde10a2 mutant D564A in complex with cAMP. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Wang, H.C, Liu, Y.D, Hou, J, Zheng, M.Y, Robinson, H. | | Deposit date: | 2007-02-12 | | Release date: | 2007-03-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | From the Cover: Structural insight into substrate specificity of phosphodiesterase 10.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1MFN
 
 | | SOLUTION NMR STRUCTURE OF LINKED CELL ATTACHMENT MODULES OF MOUSE FIBRONECTIN CONTAINING THE RGD AND SYNERGY REGIONS, 20 STRUCTURES | | Descriptor: | FIBRONECTIN | | Authors: | Copie, V, Tomita, Y, Akiyama, S.K, Aota, S, Yamada, K.M, Venable, R.M, Pastor, R.W, Krueger, S, Torchia, D.A. | | Deposit date: | 1998-01-27 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of linked cell attachment modules of mouse fibronectin containing the RGD and synergy regions: comparison with the human fibronectin crystal structure.
J.Mol.Biol., 277, 1998
|
|
4RG7
 
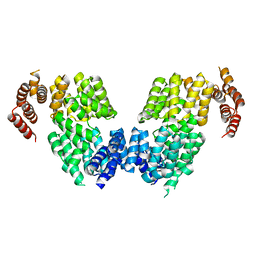 | | Crystal structure of APC3 | | Descriptor: | Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
2Y2F
 
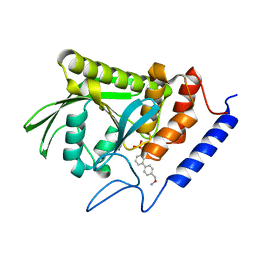 | | Crystal structure of Yersinia pestis YopH in complex with an aminooxy- containing platform compound for inhibitor design | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE YOPH, [4-[3-(DIFLUORO-PHOSPHONO-METHYL)PHENYL]PHENYL]METHOXYAZANIUM | | Authors: | Lountos, G.T, Bahta, M, Dyas, B, Ulrich, R.G, Waugh, D.S, Burke, T.R. | | Deposit date: | 2010-12-14 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Utilization of Nitrophenylphosphates and Oxime-Based Ligation for the Development of Nanomolar Affinity Inhibitors of the Yersinia Pestis Outer Protein H (Yoph) Phosphatase.
J.Med.Chem., 54, 2011
|
|
2PCS
 
 | | Crystal structure of conserved protein from Geobacillus kaustophilus | | Descriptor: | Conserved protein, UNKNOWN LIGAND | | Authors: | Bonanno, J.B, Gilmore, J, Bain, K.T, Wu, B, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved protein from Geobacillus kaustophilus
To be Published
|
|
3ZFS
 
 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
7WEK
 
 | |
2F1S
 
 | | Crystal Structure of a Viral FLIP MC159 | | Descriptor: | Viral CASP8 and FADD-like apoptosis regulator | | Authors: | Li, F.-Y, Jeffrey, P.D, Yu, J.W, Shi, Y. | | Deposit date: | 2005-11-15 | | Release date: | 2005-11-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of a Viral FLIP: INSIGHTS INTO FLIP-MEDIATED INHIBITION OF DEATH RECEPTOR SIGNALING.
J.Biol.Chem., 281, 2006
|
|
7WEJ
 
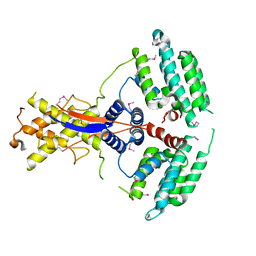 | | Crystal structure of the mouse Wdr47 NTD. | | Descriptor: | WD repeat-containing protein 47 | | Authors: | Ren, J.Q, Li, D, Feng, W. | | Deposit date: | 2021-12-23 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Intertwined Wdr47-NTD dimer recognizes a basic-helical motif in Camsap proteins for proper central-pair microtubule formation.
Cell Rep, 41, 2022
|
|
2F3V
 
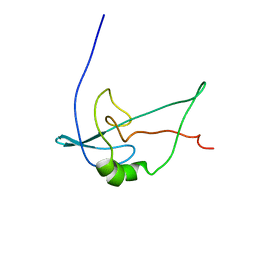 | | Solution structure of 1-110 fragment of staphylococcal nuclease with V66W mutation | | Descriptor: | Thermonuclease | | Authors: | Liu, D, Xie, T, Feng, Y, Shan, L, Ye, K, Wang, J. | | Deposit date: | 2005-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Folding stability and cooperativity of the three forms of 1-110 residues fragment of staphylococcal nuclease
Biophys.J., 92, 2007
|
|
4R7G
 
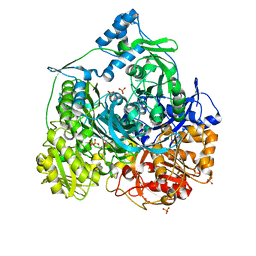 | | Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Tanwar, A.S, Sindhikara, D.J, Hirata, F, Anand, R. | | Deposit date: | 2014-08-27 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Determination of the formylglycinamide ribonucleotide amidotransferase ammonia pathway by combining 3D-RISM theory with experiment.
Acs Chem.Biol., 10, 2015
|
|
4RAG
 
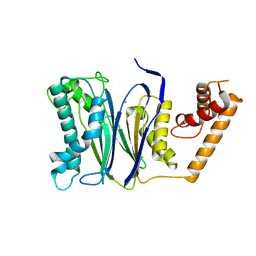 | | CRYSTAL STRUCTURE of PPC2A-D38K | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-10 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2C alpha
Sci Rep, 5, 2015
|
|
2VC8
 
 | |
2V92
 
 | | Crystal structure of the regulatory fragment of mammalian AMPK in complexes with ATP-AMP | | Descriptor: | 5'-AMP-ACTIVATED PROTEIN KINASE CATALYTIC SUBUNIT ALPHA-1, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT BETA-2, 5'-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-1, ... | | Authors: | Xiao, B, Heath, R, Saiu, P, Leiper, F.C, Leone, P, Jing, C, Walker, P.A, Haire, L, Eccleston, J.F, Davis, C.T, Martin, S.R, Carling, D, Gamblin, S.J. | | Deposit date: | 2007-08-20 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for AMP Binding to Mammalian AMP-Activated Protein Kinase
Nature, 449, 2007
|
|
5Q1E
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 5-bromo-1-{[4-(1H-tetrazol-5-yl)phenyl]methyl}-1'-(thiophene-2-sulfonyl)spiro[indole-3,4'-piperidin]-2(1H)-one, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
4B5Y
 
 | | X-ray structure of the cyan fluorescent protein mTurquoise-GL (K206A mutant) in space group C222(1) | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | von Stetten, D, Lelimousin, M, Oost, K, Noirclerc-Savoye, M, Gadella, T.W.J, Goedhart, J, Royant, A. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Influence of the H148G Mutation on Fluorescence Properties of Cyan Fluorescent Proteins
To be Published
|
|
7YO4
 
 | |
7YO0
 
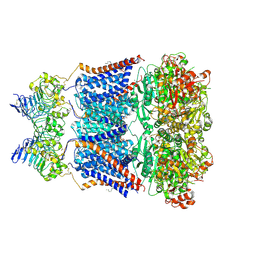 | | Cryo-EM structure of human Slo1-LRRC26 complex with Symmetry Expansion | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CALCIUM ION, Calcium-activated potassium channel subunit alpha-1, ... | | Authors: | Yamanouchi, D, Nureki, O. | | Deposit date: | 2022-08-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for dual allosteric gating modulation of Slo1-LRRC channel complex
To Be Published
|
|
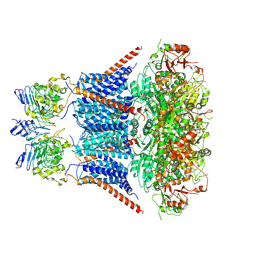
7YNZ
 
 | |
2VAI
 
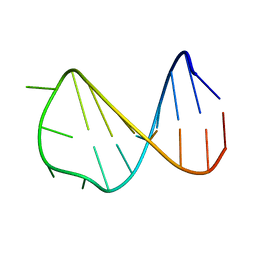 | | Solution structure of a B-DNA hairpin at high pressure | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural change in a B-DNA helix with hydrostatic pressure.
Nucleic Acids Res., 36, 2008
|
|
2QKR
 
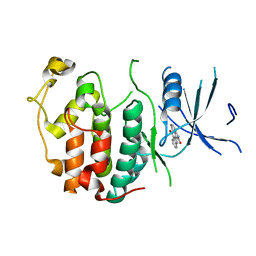 | | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3'-monoxime bound | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, Cdc2-like CDK2/CDC28 like protein kinase | | Authors: | Wernimont, A.K, Dong, A, Lew, J, Lin, Y.H, Hassanali, A, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-07-11 | | Release date: | 2007-09-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryptosporidium parvum cyclin-dependent kinase cgd5_2510 with indirubin 3'-monoxime bound.
To be Published
|
|
1MV2
 
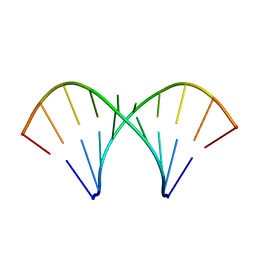 | | The tandem, Face-to-Face AP Pairs in 5'(rGGCAPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*AP*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
1MV6
 
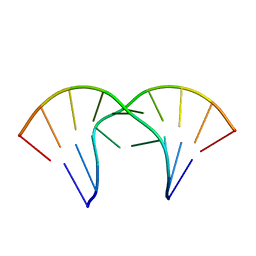 | | The tandem, Sheared PP Pairs in 5'(rGGCPPGCCU)2 | | Descriptor: | 5'-R(*GP*GP*CP*(P5P)P*(P5P)P*GP*CP*CP*U)-3' | | Authors: | Znosko, B.M, Burkard, M.E, Krugh, T.R, Turner, D.H. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition in Purine-Rich Internal Loops: Thermodynamic, Structural, and Dynamic Consequences of Purine for Adenine Substitutions in 5'(rGGCAAGCCU)2
Biochemistry, 41, 2002
|
|
2AQA
 
 | |
