4TTR
 
 | |
1F5H
 
 | |
7CQB
 
 | |
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | 著者 | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | 登録日 | 2015-01-19 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6I6S
 
 | | Circular permutant of ribosomal protein S6, adding 9aa to C terminal of P68-69, L75A mutant | | 分子名称: | 30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6,30S ribosomal protein S6, POTASSIUM ION, SODIUM ION | | 著者 | Wang, H, Logan, D.T, Oliveberg, M. | | 登録日 | 2018-11-15 | | 公開日 | 2019-11-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Exposing the distinctive modular behavior of beta-strands and alpha-helices in folded proteins.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7YNN
 
 | | ThT-bound alpha-synuclein fibrils conformation 2 | | 分子名称: | 2-[4-(dimethylamino)phenyl]-3,6-dimethyl-1,3-benzothiazol-3-ium, Alpha-synuclein | | 著者 | Tao, Y.Q, Zhao, Q.Y, Liu, C, Li, D. | | 登録日 | 2022-07-31 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | ThT-bound alpha-synuclein fibrils conformation 2
To Be Published
|
|
6ZJ4
 
 | | apo-Trehalose transferase (apo-TreT) from Thermoproteus uzoniensis | | 分子名称: | THIOCYANATE ION, Trehalose phosphorylase/synthase | | 著者 | Bento, I, Mestrom, L, Marsden, S.R, van der Eijk, H, Laustsen, J.U, Jeffries, C.M, Svergun, D.I, Hagedoorn, P.-H, Hanefeld, U. | | 登録日 | 2020-06-27 | | 公開日 | 2020-09-30 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Anomeric Selectivity of Trehalose Transferase with Rare l-Sugars.
Acs Catalysis, 10, 2020
|
|
1RE6
 
 | | Localisation of Dynein Light Chains 1 and 2 and their Pro-apoptotic Ligands | | 分子名称: | dynein light chain 2 | | 著者 | Day, C.L, Puthalakath, H, Skea, G, Strasser, A, Barsukov, I, Lian, L.Y, Huang, D.C, Hinds, M.G. | | 登録日 | 2003-11-06 | | 公開日 | 2004-03-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Localization of dynein light chains 1 and 2 and their pro-apoptotic ligands.
Biochem.J., 377, 2004
|
|
7YNO
 
 | |
6WQZ
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | 分子名称: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | 著者 | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | 登録日 | 2020-04-29 | | 公開日 | 2020-07-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
5HM9
 
 | | Crystal structure of MamO protease domain from Magnetospirillum magneticum (apo form) | | 分子名称: | MamO protease domain, poly(UNK) | | 著者 | Hershey, D.M, Ren, X, Hurley, J.H, Komeili, A. | | 登録日 | 2016-01-15 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Plos Biol., 14, 2016
|
|
6NNQ
 
 | |
9CMP
 
 | | Structure of human Argonaute2-guide-target complex in a fully paired, slicing-competent conformation | | 分子名称: | MAGNESIUM ION, Protein argonaute-2, RNA (5'-R(*CP*AP*AP*CP*AP*AP*AP*AP*UP*CP*AP*CP*UP*AP*GP*UP*CP*UP*UP*CP*CP*A)-3'), ... | | 著者 | Mohamed, A.A, Wang, P.Y, Bartel, D.P, Vos, S.M. | | 登録日 | 2024-07-15 | | 公開日 | 2024-12-11 | | 最終更新日 | 2025-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | The structural basis for RNA slicing by human Argonaute2.
Cell Rep, 44, 2025
|
|
6ZLY
 
 | |
1E10
 
 | | [2Fe-2S]-Ferredoxin from Halobacterium salinarum | | 分子名称: | ACETYL GROUP, FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | 著者 | Marg, B.-L, Schweimer, K, Oesterhelt, D, Roesch, P, Sticht, H. | | 登録日 | 2000-04-12 | | 公開日 | 2001-04-09 | | 最終更新日 | 2025-04-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Two-Alpha-Helix Extra Domain Mediates the Halophilic Character of a Plant-Type Ferredoxin from Halophilic Archaea.
Biochemistry, 44, 2005
|
|
8BSC
 
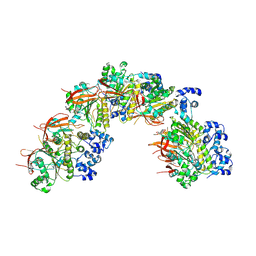 | |
9CJC
 
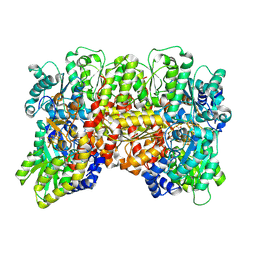 | |
6NSK
 
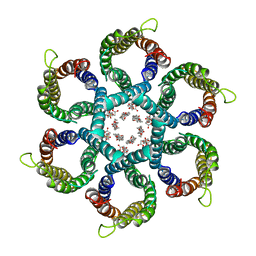 | | CryoEM structure of Helicobacter pylori urea channel in open state. | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, Acid-activated urea channel | | 著者 | Cui, Y.X, Zhou, K, Strugatsky, D, Wen, Y, Sachs, G, Munson, K, Zhou, Z.H. | | 登録日 | 2019-01-24 | | 公開日 | 2019-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | pH-dependent gating mechanism of theHelicobacter pyloriurea channel revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6WI5
 
 | |
4RSD
 
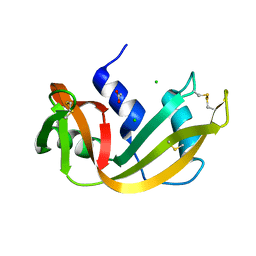 | | STRUCTURE OF THE D121A VARIANT OF RIBONUCLEASE A | | 分子名称: | ACETATE ION, CHLORIDE ION, RIBONUCLEASE A | | 著者 | Schultz, L.W, Quirk, D.J, Raines, R.T. | | 登録日 | 1998-02-05 | | 公開日 | 1998-07-15 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | His...Asp catalytic dyad of ribonuclease A: structure and function of the wild-type, D121N, and D121A enzymes.
Biochemistry, 37, 1998
|
|
7MYV
 
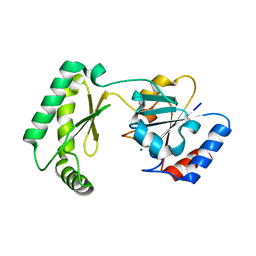 | |
7N05
 
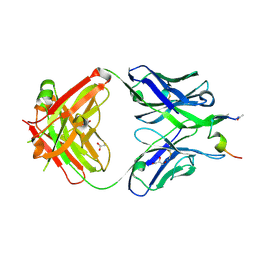 | |
5R5U
 
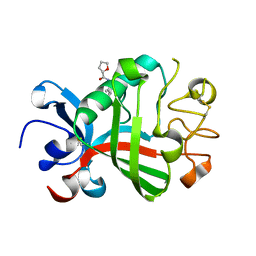 | | PanDDA analysis group deposition -- Crystal Structure of FIBRINOGEN-LIKE GLOBE DOMAIN OF HUMAN TENASCIN-C in complex with Z1545312521 | | 分子名称: | (2S)-N-(3-chloro-2-methylphenyl)oxolane-2-carboxamide, Tenascin C (Hexabrachion), isoform CRA_a | | 著者 | Coker, J.A, Bezerra, G.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Yue, W.W, Marsden, B.D. | | 登録日 | 2020-02-28 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | PanDDA analysis group deposition
To Be Published
|
|
8C70
 
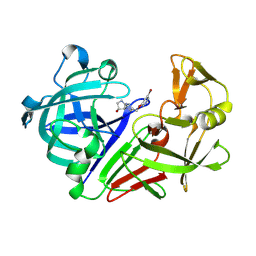 | | Pyrrolidine fragment 1 bound to endothiapepsin | | 分子名称: | (3~{R},4~{R})-4-[4-[(5-bromanylpyridin-3-yl)oxymethyl]-1,2,3-triazol-1-yl]pyrrolidin-3-ol, Endothiapepsin | | 著者 | Wiese, J.N, Buehrmann, M, Mueller, M.P, Rauh, D. | | 登録日 | 2023-01-12 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Fragtory: Pharmacophore-Focused Design, Synthesis, and Evaluation of an sp 3 -Enriched Fragment Library.
J.Med.Chem., 66, 2023
|
|
1EF4
 
 | | SOLUTION STRUCTURE OF THE ESSENTIAL RNA POLYMERASE SUBUNIT RPB10 FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | 分子名称: | DNA-DIRECTED RNA POLYMERASE, ZINC ION | | 著者 | Mackereth, C.D, Arrowsmith, C.H, Edwards, A.M, Mcintosh, L.P, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2000-02-07 | | 公開日 | 2000-06-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Zinc-bundle structure of the essential RNA polymerase subunit RPB10 from Methanobacterium thermoautotrophicum.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
