7R06
 
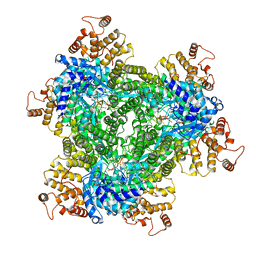 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3') | | Authors: | Figiel, M, Nowotny, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7R07
 
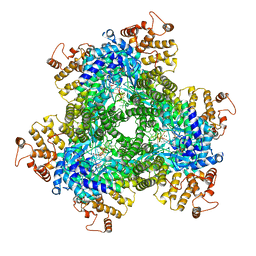 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis | | Descriptor: | AbiK, DNA (5'-D(*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*C)-3'), MAGNESIUM ION | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-01 | | Release date: | 2022-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
7PIK
 
 | | Cryo-EM structure of E. coli TnsB in complex with right end fragment of Tn7 transposon | | Descriptor: | Right end fragment of Tn7 transposon, Transposon Tn7 transposition protein TnsB | | Authors: | Kaczmarska, Z, Czarnocki-Cieciura, M, Rawski, M, Nowotny, M. | | Deposit date: | 2021-08-20 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural basis of transposon end recognition explains central features of Tn7 transposition systems.
Mol.Cell, 82, 2022
|
|
8A8J
 
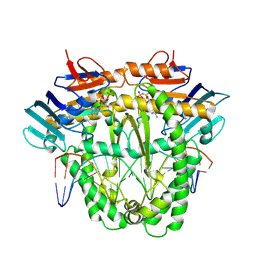 | | Complex of RecF and DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A93
 
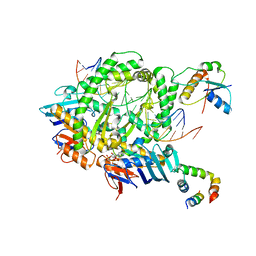 | | Complex of RecF-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA replication and repair protein RecF, MAGNESIUM ION, Oligo1, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-06-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AB0
 
 | | Complex of RecO-RecR-DNA from Thermus thermophilus. | | Descriptor: | DNA repair protein RecO, Oligo1, Oligo2, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (6.09 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z0Z
 
 | | Abortive infection DNA polymerase AbiK from Lactococcus lactis, Y44F variant | | Descriptor: | AbiK | | Authors: | Figiel, M, Gapinska, M, Czarnocki-Cieciura, M, Zajko, W, Nowotny, M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Mechanism of protein-primed template-independent DNA synthesis by Abi polymerases.
Nucleic Acids Res., 50, 2022
|
|
9HFT
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with 5mdCTP bound in the active site | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION, [[(2~{R},3~{S},5~{R})-5-(4-azanyl-5-methyl-2-oxidanylidene-pyrimidin-1-yl)-3-oxidanyl-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFR
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFS
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): hexamer with dCTP bound in the active site | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9HFQ
 
 | | Cryo-EM structure of human CDADC1 inactive mutant (E400A): trimer without a ligand | | Descriptor: | Cytidine and dCMP deaminase domain-containing protein 1, ZINC ION | | Authors: | Slyvka, A, Rathore, I, Yang, R, Kanai, T, Lountos, G, Wang, Z, Skowronek, K, Czarnocki-Cieciura, M, Wlodawer, A, Bochtler, M. | | Deposit date: | 2024-11-18 | | Release date: | 2025-04-30 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Activity and structure of human (d)CTP deaminase CDADC1.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8BPR
 
 | | Complex of RecF-RecO-RecR-DNA from Thermus thermophilus (low resolution reconstruction). | | Descriptor: | DNA repair protein RecO, DNA replication and repair protein RecF, MAGNESIUM ION, ... | | Authors: | Nirwal, S, Czarnocki-Cieciura, M, Chaudhary, A, Zajko, W, Skowronek, K, Chamera, S, Figiel, M, Nowotny, M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-04-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Mechanism of RecF-RecO-RecR cooperation in bacterial homologous recombination.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BDC
 
 | | Human apo TRPM8 in a closed state (composite map) | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Palchevskyi, S, Czarnocki-Cieciura, M, Vistoli, G, Gervasoni, S, Nowak, E, Beccari, A.R, Nowotny, M, Talarico, C. | | Deposit date: | 2022-10-19 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structure of human TRPM8 channel.
Commun Biol, 6, 2023
|
|
7O24
 
 | |
9I8V
 
 | | Cryo-EM structure of the Danio rerio tRNA ligase complex | | Descriptor: | ATP-dependent RNA helicase, Ashwin, Family with sequence similarity 98, ... | | Authors: | Chamera, S, Zajko, W, Czarnocki-Cieciura, M, Jaciuk, M, Koziej, L, Nowak, J, Wycisk, K, Sroka, M, Chramiec-Glabik, A, Smietanski, M, Golebiowski, F, Warminski, M, Jemielity, J, Glatt, S, Nowotny, M. | | Deposit date: | 2025-02-06 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural and biochemical characterization of the 3'-5' tRNA splicing ligases.
J.Biol.Chem., 301, 2025
|
|
9FRX
 
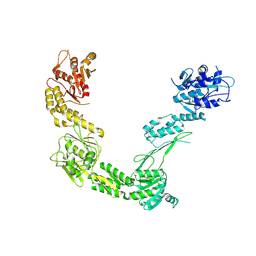 | | Porcine Retinol-Binding Protein 3 (RBP3) | | Descriptor: | Retinol binding protein 3 | | Authors: | Kaushik, V, Gessa, L, Kumar, N, Pinkas, M, Czarnocki-Cieciura, M, Palczewski, K, Novacek, J, Fernandes, H. | | Deposit date: | 2024-06-19 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | CryoEM structure and small-angle X-ray scattering analyses of porcine retinol-binding protein 3.
Open Biology, 15, 2025
|
|
