2W8C
 
 | |
2W88
 
 | |
3CVB
 
 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (I) ION, Plastocyanin | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
3CVC
 
 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (II) ION, MAGNESIUM ION, Plastocyanin | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
3CVD
 
 | | Regulation of Protein Function: Crystal Packing Interfaces and Conformational Dimerization | | Descriptor: | COPPER (I) ION, Plastocyanin, ZINC ION | | Authors: | Crowley, P.B, Matias, P.M, Mi, H, Firbank, S.J, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Regulation of protein function: crystal packing interfaces and conformational dimerization.
Biochemistry, 47, 2008
|
|
4N0J
 
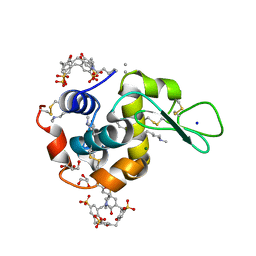 | | Crystal structure of dimethyllysine hen egg-white lysozyme in complex with sclx4 at 1.9 A resolution | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | McGovern, R.E, Crowley, P.B. | | Deposit date: | 2013-10-02 | | Release date: | 2014-11-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural study of a small molecule receptor bound to dimethyllysine in lysozyme.
Chem Sci, 6, 2015
|
|
3TYI
 
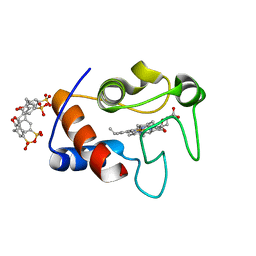 | | Crystal Structure of Cytochrome c - p-Sulfonatocalix[4]arene Complexes | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mc Govern, R.E, Fernandes, H, Khan, A.R, Crowley, P.B. | | Deposit date: | 2011-09-26 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Protein camouflage in cytochrome c-calixarene complexes.
Nat Chem, 4, 2012
|
|
5T8W
 
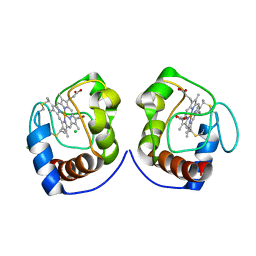 | |
6EGY
 
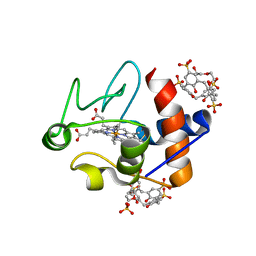 | | Crystal structure of cytochrome c in complex with mono-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SULFATE ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
6EGZ
 
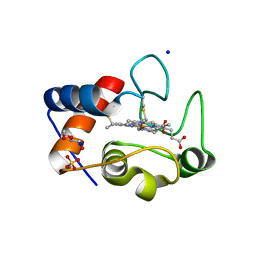 | | Crystal structure of cytochrome c in complex with di-PEGylated sulfonatocalix[4]arene | | Descriptor: | Cytochrome c iso-1, HEME C, SODIUM ION, ... | | Authors: | Mummidivarapu, V.V.S, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-09-12 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Noncovalent PEGylation via Sulfonatocalix[4]arene-A Crystallographic Proof.
Bioconjug.Chem., 29, 2018
|
|
4YE1
 
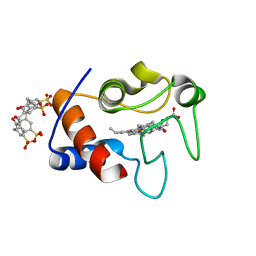 | | A cytochrome c plus calixarene structure - alternative ligand binding mode | | Descriptor: | 25,26,27,28-tetrahydroxypentacyclo[19.3.1.1~3,7~.1~9,13~.1~15,19~]octacosa-1(25),3(28),4,6,9(27),10,12,15(26),16,18,21,23-dodecaene-5,11,17,23-tetrasulfonic acid, Cytochrome c iso-1, GLYCEROL, ... | | Authors: | Mallon, M.M, McGovern, R.E, McCarty, A.A, Crowley, P.B. | | Deposit date: | 2015-02-23 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A cytochrome c-calixarene structure
To Be Published
|
|
8Q6C
 
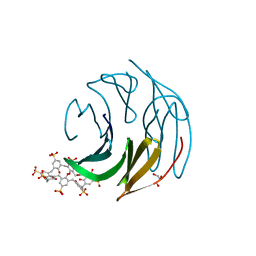 | | The RSL-D32N - sulfonato-calix[8]arene complex, P63 form, acetate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
8Q6B
 
 | | The RSL-D32N - sulfonato-calix[8]arene complex, I23 form, citrate pH 4.0, obtained by cross-seeding | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
8Q6A
 
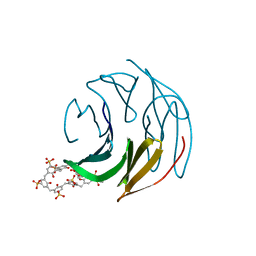 | | The RSL-D32N - sulfonato-calix[8]arene complex, I213 form, citrate pH 4.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Flood, R.J, Crowley, P.B. | | Deposit date: | 2023-08-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Supramolecular Synthons in Protein-Ligand Frameworks.
Cryst.Growth Des., 24, 2024
|
|
6Y0J
 
 | |
5NCV
 
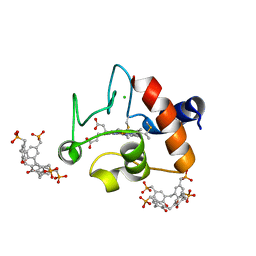 | | Crystal Structure of Cytochrome c in complex with p-Methylphosphonatocalix[4]arene | | Descriptor: | CHLORIDE ION, Cytochrome c iso-1, HEME C, ... | | Authors: | Alex, J.M, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Phosphonated Calixarene as a ""Molecular Glue"" for Protein Crystallization
Cryst.Growth Des., 18, 2018
|
|
5LYC
 
 | |
8CF6
 
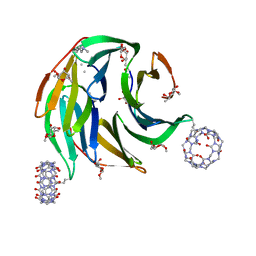 | |
8CF7
 
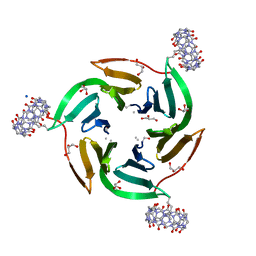 | |
8C9Z
 
 | | The RSL - sulfonato-calix[8]arene complex, H32 form, citrate pH 6.0 | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, beta-D-fructopyranose, ... | | Authors: | Mockler, N.M, Ramberg, K, Crowley, P.B. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Protein-macrocycle polymorphism: crystal form IV of the Ralstonia solanacearum lectin-sulfonato-calix[8]arene complex.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
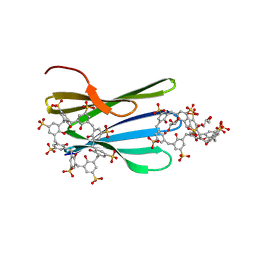
8C9Y
 
 | | The MK-RSL - sulfonato-calix[8]arene complex, H32 form | | Descriptor: | GLYCEROL, Putative fucose-binding lectin protein, beta-D-fructopyranose, ... | | Authors: | Mockler, N.M, Ramberg, K, Crowley, P.B. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Protein-macrocycle polymorphism: crystal form IV of the Ralstonia solanacearum lectin-sulfonato-calix[8]arene complex.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2JKW
 
 | | Pseudoazurin M16F | | Descriptor: | COPPER (II) ION, PSEUDOAZURIN | | Authors: | Yanagisawa, S, Crowley, P.B, Firbank, S.J, Lawler, A.T, Hunter, D.M, McFarlane, W, Li, C, Kohzuma, T, Banfield, M.J, Dennison, C. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pi-Interaction Tuning of the Active Site Properties of Metalloproteins.
J.Am.Chem.Soc., 130, 2008
|
|
7P2H
 
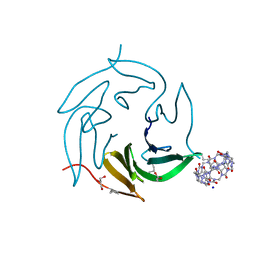 | | Dimethylated fusion protein of RSL and mussel adhesion peptide (Mefp) in complex with cucurbit[7]uril, H3 sheet assembly | | Descriptor: | Fucose-binding lectin protein, GLYCEROL, SODIUM ION, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
7P2I
 
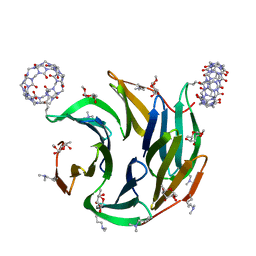 | | Dimethylated fusion protein of RSL and nucleoporin peptide (Nup) in complex with cucurbit[7]uril, F432 cage assembly | | Descriptor: | Fucose-binding lectin protein, cucurbit[7]uril, methyl alpha-L-fucopyranoside | | Authors: | Guagnini, F, Ramberg, K, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
7P2J
 
 | | Dimethylated fusion protein of RSL and trimeric coiled coil (4dzn) in complex with cucurbit[7]uril, H3 sheet assembly | | Descriptor: | Fucose-binding lectin protein, SODIUM ION, beta-D-mannopyranose, ... | | Authors: | Ramberg, K, Engilberge, S, Crowley, P.B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Segregated Protein-Cucurbit[7]uril Crystalline Architectures via Modulatory Peptide Tectons.
Chemistry, 27, 2021
|
|
