4V84
 
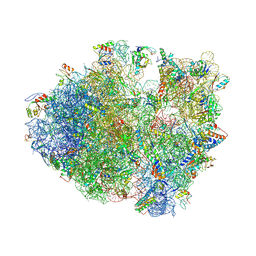 | | Crystal structure of a complex containing domain 3 of CrPV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7JJU
 
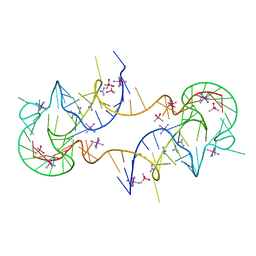 | | Crystal structure of en exoribonuclease-resistant RNA (xrRNA) from Potato leafroll virus (PLRV) | | Descriptor: | CACODYLATE ION, Guanidinium, IRIDIUM HEXAMMINE ION, ... | | Authors: | Steckelberg, A.-L, Vicens, Q, Auffinger, P, Costantino, D.C, Nix, J.C, Kieft, J.S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | The crystal structure of a Polerovirus exoribonuclease-resistant RNA shows how diverse sequences are integrated into a conserved fold.
Rna, 26, 2020
|
|
6MJ0
 
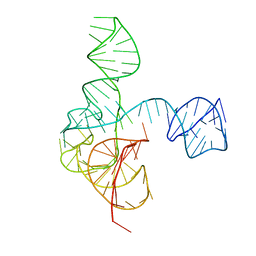 | | Crystal structure of the complete turnip yellow mosaic virus 3'UTR | | Descriptor: | RNA (101-MER) | | Authors: | Hartwick, E.W, Costantino, D.A, MacFadden, A, Nix, J.C, Tian, S, Das, R, Kieft, J.S. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Ribosome-induced RNA conformational changes in a viral 3'-UTR sense and regulate translation levels.
Nat Commun, 9, 2018
|
|
4V83
 
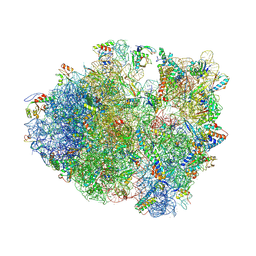 | | Crystal structure of a complex containing domain 3 from the PSIV IGR IRES RNA bound to the 70S ribosome. | | Descriptor: | 23S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Zhu, J, Korostelev, A, Costantino, D, Noller, H.F, Kieft, J.S. | | Deposit date: | 2010-12-13 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2IL9
 
 | |
5TPY
 
 | | Crystal structure of an exonuclease resistant RNA from Zika virus | | Descriptor: | HEXANE-1,6-DIOL, MAGNESIUM ION, RNA (71-MER) | | Authors: | Akiyama, B.M, Laurence, H.M, Massey, A.R, Costantino, D.A, Xie, X, Yang, Y, Shi, P.-Y, Nix, J.C, Beckham, J.D, Kieft, J.S. | | Deposit date: | 2016-10-21 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease.
Science, 354, 2016
|
|
6D3P
 
 | | Crystal structure of an exoribonuclease-resistant RNA from Sweet clover necrotic mosaic virus (SCNMV) | | Descriptor: | IRIDIUM HEXAMMINE ION, RNA (45-MER) | | Authors: | Steckelberg, A.-L, Akiyama, B.M, Costantino, D.A, Sit, T.L, Nix, J.C, Kieft, J.S. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A folded viral noncoding RNA blocks host cell exoribonucleases through a conformationally dynamic RNA structure.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3B31
 
 | | Crystal structure of domain III of the Cricket Paralysis Virus IRES RNA | | Descriptor: | IRIDIUM HEXAMMINE ION, PHOSPHATE ION, RNA (29-MER), ... | | Authors: | Kieft, J.S, Costantino, D.A, Pfingsten, J.S, Rambo, R.P. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | tRNA-mRNA mimicry drives translation initiation from a viral IRES.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3MJB
 
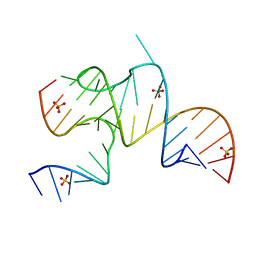 | | Cricket Paralysis Virus IGR IRES Domain 3 RNA bound to sulfate | | Descriptor: | Domain 3 of the cricket paralysis virus intergenic region IRES RNA, RNA (5'-R(P*UP*AP*AP*GP*AP*AP*AP*UP*UP*UP*AP*CP*CP*U)-3'), SULFATE ION | | Authors: | Kieft, J.S, Golden, B.L, Costantino, D.A, Chase, E. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification and characterization of anion binding sites in RNA.
Rna, 16, 2010
|
|
6DVK
 
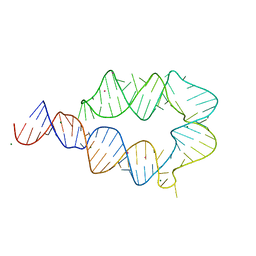 | | Computationally designed mini tetraloop-tetraloop receptor by the RNAMake program - construct 6 (miniTTR 6) | | Descriptor: | COBALT (II) ION, MAGNESIUM ION, RNA (95-MER) | | Authors: | Eiler, D.R, Yesselman, J.D, Costantino, D.A, Das, R, Kieft, J.S. | | Deposit date: | 2018-06-24 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Computational design of three-dimensional RNA structure and function.
Nat Nanotechnol, 14, 2019
|
|
3MJA
 
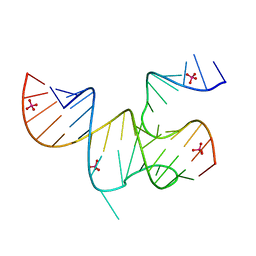 | | Cricket Paralysis Virus IGR IRES Domain 3 RNA bound to selenate, structure #2 | | Descriptor: | Domain 3 of the cricket paralysis virus intergenic region IRES RNA, RNA (5'-R(P*UP*AP*AP*GP*AP*AP*AP*UP*UP*UP*AP*CP*CP*U)-3'), SELENATE ION | | Authors: | Kieft, J.S, Golden, B.L, Costantino, D.A, Chase, E. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification and characterization of anion binding sites in RNA.
Rna, 16, 2010
|
|
3MJ3
 
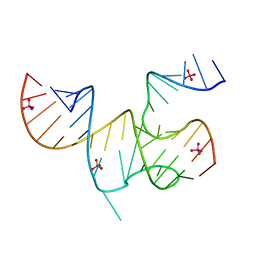 | | Cricket Paralysis Virus IGR IRES Domain 3 RNA bound to selenate | | Descriptor: | Domain 3 of the cricket paralysis virus intergenic region IRES RNA, RNA (5'-R(P*UP*AP*AP*GP*AP*AP*AP*UP*UP*UP*AP*CP*CP*U)-3'), SELENATE ION | | Authors: | Kieft, J.S, Chase, E, Costantino, D.A, Golden, B.L. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification and characterization of anion binding sites in RNA.
Rna, 16, 2010
|
|
4P5J
 
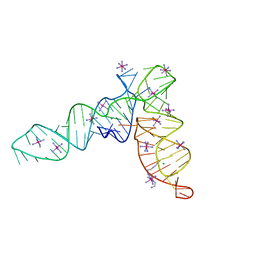 | | Crystal structure of the tRNA-like structure from Turnip Yellow Mosaic Virus (TYMV), a tRNA mimicking RNA | | Descriptor: | IRIDIUM HEXAMMINE ION, MAGNESIUM ION, SPERMINE, ... | | Authors: | Colussi, T.M, Costantino, D.A, Hammond, J.A, Ruehle, G.M, Nix, J.C, Kieft, J.S. | | Deposit date: | 2014-03-17 | | Release date: | 2014-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9912 Å) | | Cite: | The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA.
Nature, 511, 2014
|
|
4PQV
 
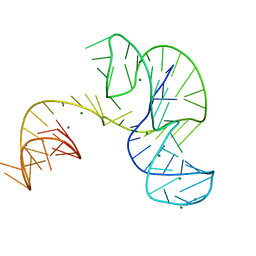 | | Crystal structure of an Xrn1-resistant RNA from the 3' untranslated region of a flavivirus (Murray Valley Encephalitis virus) | | Descriptor: | MAGNESIUM ION, XRN1-resistant flaviviral RNA | | Authors: | Chapman, E.G, Costantino, D.A, Rabe, J.L, Moon, S.L, Wilusz, J, Nix, J.C, Kieft, J.S. | | Deposit date: | 2014-03-04 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.463 Å) | | Cite: | The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production.
Science, 344, 2014
|
|
