1NLA
 
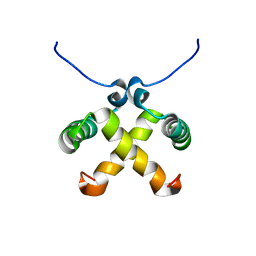 | | Solution Structure of Switch Arc, a Mutant with 3(10) Helices Replacing a Wild-Type Beta-Ribbon | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon
J.Mol.Biol., 326, 2003
|
|
1QTG
 
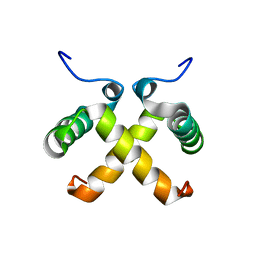 | | AVERAGED NMR MODEL OF SWITCH ARC, A DOUBLE MUTANT OF ARC REPRESSOR | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H.J, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 1999-06-27 | | Release date: | 1999-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Evolution of a protein fold in vitro.
Science, 284, 1999
|
|
2A63
 
 | |
5W8Z
 
 | |
3BD1
 
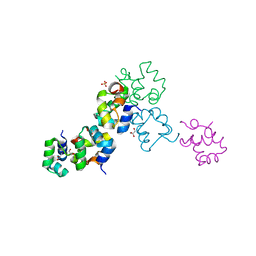 | | Structure of the Cro protein from putative prophage element Xfaso 1 in Xylella fastidiosa strain Ann-1 | | Descriptor: | CHLORIDE ION, Cro protein, GLYCEROL, ... | | Authors: | Hall, B.M, Roberts, S.A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2007-11-13 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of
Cro proteins with 40% sequence identity but different folds
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4Q6X
 
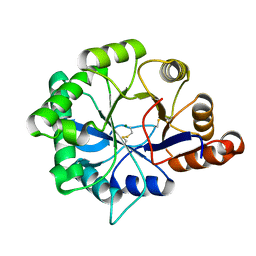 | | Structure of phospholipase D Beta1B1i from Sicarius terrosus venom at 2.14 A resolution | | Descriptor: | MAGNESIUM ION, Phospholipase D StSicTox-betaIC1 | | Authors: | Lajoie, D.M, Roberts, S.A, Zobel-Thropp, P.A, Binford, G.J, Cordes, M.H. | | Deposit date: | 2014-04-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Variable Substrate Preference among Phospholipase D Toxins from Sicariid Spiders.
J.Biol.Chem., 290, 2015
|
|
2HIN
 
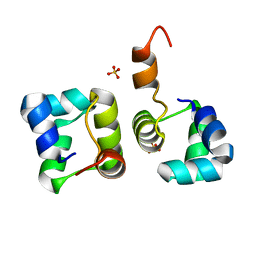 | | Structure of N15 Cro at 1.05 A: an ortholog of lambda Cro with a completely different but equally effective dimerization mechanism | | Descriptor: | Repressor protein, SULFATE ION | | Authors: | Dubrava, M.S, Ingram, W.M, Roberts, S.A, Weichsel, A, Montfort, W.R, Cordes, M.H. | | Deposit date: | 2006-06-29 | | Release date: | 2007-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | N15 Cro and lambda Cro: orthologous DNA-binding domains with completely different but equally effective homodimer interfaces.
Protein Sci., 17, 2008
|
|
5W8Y
 
 | |
2ECS
 
 | | Lambda Cro mutant Q27P/A29S/K32Q at 1.4 A in space group C2 | | Descriptor: | ACETATE ION, CHLORIDE ION, LITHIUM ION, ... | | Authors: | Hall, B.M, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
2OVG
 
 | | Lambda Cro Q27P/A29S/K32Q triple mutant at 1.35 A in space group P3221 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Phage lambda Cro, SULFATE ION | | Authors: | Hall, B.M, Heroux, A, Roberts, S.A, Cordes, M.H. | | Deposit date: | 2007-02-13 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Two structures of a lambda Cro variant highlight dimer flexibility but disfavor major dimer distortions upon specific binding of cognate DNA.
J.Mol.Biol., 375, 2008
|
|
1RZS
 
 | |
2PIJ
 
 | | Structure of the Cro protein from prophage Pfl 6 in Pseudomonas fluorescens Pf-5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BICARBONATE ION, Prophage Pfl 6 Cro, ... | | Authors: | Roessler, C.G, Roberts, S.A, Montfort, W.R, Cordes, M.H.J. | | Deposit date: | 2007-04-13 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
6ON0
 
 | | STRUCTURE OF N15 CRO COMPLEXED WITH CONSENSUS OPERATOR DNA | | Descriptor: | DNA (5'-D(*TP*TP*TP*AP*TP*AP*GP*CP*TP*AP*GP*CP*TP*AP*TP*AP*A)-3'), Gp39 | | Authors: | Hall, B.M, Roberts, S.A, Cordes, M.H.J. | | Deposit date: | 2019-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extreme divergence between one-to-one orthologs: the structure of N15 Cro bound to operator DNA and its relationship to the lambda Cro complex.
Nucleic Acids Res., 47, 2019
|
|
