2QE0
 
 | | Thioacylenzyme Intermediate of GAPN from S. Mutans, New Data Integration and Refinement. | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Corbier, C, Didierjean, C, Bricogne, G, Branlant, G, D'Ambrosio, K, Vonrhein, C. | | Deposit date: | 2007-06-22 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The First Crystal Structure of a Thioacylenzyme Intermediate in the ALDH Family: New Coenzyme Conformation and Relevance to Catalysis
Biochemistry, 45, 2006
|
|
2ESD
 
 | | Crystal Structure of thioacylenzyme intermediate of an Nadp Dependent Aldehyde Dehydrogenase | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase | | Authors: | D'Ambrosio, K, Didierjean, C, Benedetti, E, Aubry, A, Corbier, C. | | Deposit date: | 2005-10-26 | | Release date: | 2006-05-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis
Biochemistry, 45, 2006
|
|
2XF8
 
 | | Structure of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli in complex with a NAD cofactor analog (3-Chloroacetyl adenine pyridine dinucleotide) and sulfate anion | | Descriptor: | 3-(CHLOROACETYL) PYRIDINE ADENINE DINUCLEOTIDE, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, SULFATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-05-20 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
3CMC
 
 | | Thioacylenzyme intermediate of Bacillus stearothermophilus phosphorylating GAPDH | | Descriptor: | 1,2-ETHANEDIOL, GLYCERALDEHYDE-3-PHOSPHATE, GLYCEROL, ... | | Authors: | Moniot, S, Vonrhein, C, Bricogne, G, Didierjean, C, Corbier, C. | | Deposit date: | 2008-03-21 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Trapping of the Thioacylglyceraldehyde-3-phosphate Dehydrogenase Intermediate from Bacillus stearothermophilus: DIRECT EVIDENCE FOR A FLIP-FLOP MECHANISM
J.Biol.Chem., 283, 2008
|
|
1T90
 
 | | Crystal structure of methylmalonate semialdehyde dehydrogenase from Bacillus subtilis | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Probable methylmalonate-semialdehyde dehydrogenase | | Authors: | Dubourg, H, Didierjean, C, Stines-Chaumeil, C, Talfournier, F, Branlant, G, Aubry, A, Corbier, C. | | Deposit date: | 2004-05-14 | | Release date: | 2006-01-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure analysis of Methylmalonate-Semialdehyde Dehydrogenase from Bacillus subtilis.
To be published
|
|
1GAE
 
 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
3FZ9
 
 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione | | Descriptor: | GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
2ID2
 
 | | GAPN T244S mutant X-ray structure at 2.5 A | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Pailot, A, D'Ambrosio, K, Corbier, C, Talfournier, F, Branlant, G. | | Deposit date: | 2006-09-14 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Invariant Thr(244) is essential for the efficient acylation step of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Streptococcus mutans.
Biochem.J., 400, 2006
|
|
3FZA
 
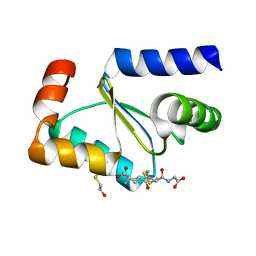 | | Crystal structure of poplar glutaredoxin S12 in complex with glutathione and beta-mercaptoethanol | | Descriptor: | BETA-MERCAPTOETHANOL, GLUTATHIONE, Glutaredoxin | | Authors: | Didierjean, C, Corbier, C, Koh, C.S, Rouhier, N, Jacquot, J.P. | | Deposit date: | 2009-01-24 | | Release date: | 2009-02-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationship of the chloroplastic glutaredoxin S12 with an atypical WCSYS active site.
J.Biol.Chem., 284, 2009
|
|
2P5R
 
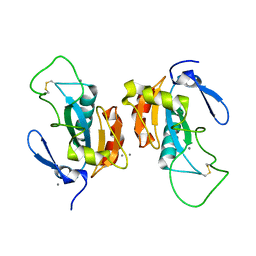 | | Crystal structure of the poplar glutathione peroxidase 5 in the oxidized form | | Descriptor: | CALCIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
2P5Q
 
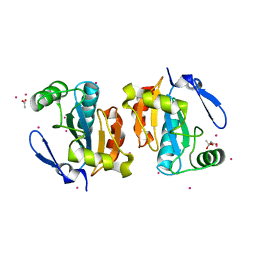 | | Crystal structure of the poplar glutathione peroxidase 5 in the reduced form | | Descriptor: | ACETATE ION, CADMIUM ION, Glutathione peroxidase 5 | | Authors: | Koh, C.S, Didierjean, C, Navrot, N, Panjikar, S, Mulliert, G, Rouhier, N, Jacquot, J.-P, Aubry, A, Shawkataly, O, Corbier, C. | | Deposit date: | 2007-03-16 | | Release date: | 2007-07-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of a Poplar Thioredoxin Peroxidase that Exhibits the Structure of Glutathione Peroxidases: Insights into Redox-driven Conformational Changes.
J.Mol.Biol., 370, 2007
|
|
1GAD
 
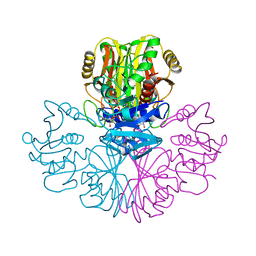 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
3D21
 
 | | Crystal structure of a poplar wild-type thioredoxin h, PtTrxh4 | | Descriptor: | Thioredoxin H-type | | Authors: | Koh, C.S, Didierjean, C, Corbier, C, Rouhier, N, Jacquot, J.P, Gelhaye, E. | | Deposit date: | 2008-05-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | An Atypical Catalytic Mechanism Involving Three Cysteines of Thioredoxin.
J.Biol.Chem., 283, 2008
|
|
3D22
 
 | | Crystal structure of a poplar thioredoxin h mutant, PtTrxh4C61S | | Descriptor: | PHOSPHATE ION, Thioredoxin H-type | | Authors: | Koh, C.S, Didierjean, C, Corbier, C, Rouhier, N, Jacquot, J.P, Gelhaye, E. | | Deposit date: | 2008-05-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Atypical Catalytic Mechanism Involving Three Cysteines of Thioredoxin.
J.Biol.Chem., 283, 2008
|
|
2X5K
 
 | | Structure of an active site mutant of the D-Erythrose-4-Phosphate Dehydrogenase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, ... | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
2X5J
 
 | | Crystal structure of the Apoform of the D-Erythrose-4-phosphate dehydrogenase from E. coli | | Descriptor: | D-ERYTHROSE-4-PHOSPHATE DEHYDROGENASE, PHOSPHATE ION | | Authors: | Moniot, S, Didierjean, C, Boschi-Muller, S, Branlant, G, Corbier, C. | | Deposit date: | 2010-02-09 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Erythrose-4- Phosphate Dehydrogenase from Escherichia Coli: Peculiar Features When Compared to Phosphorylating Gapdhs
To be Published
|
|
1TP9
 
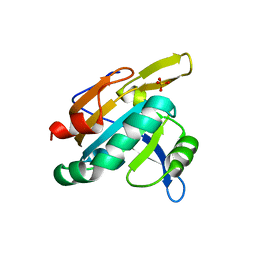 | | PRX D (type II) from Populus tremula | | Descriptor: | SULFATE ION, peroxiredoxin | | Authors: | Echalier, A, Trivelli, X, Corbier, C, Rouhier, N, Walker, O, Tsan, P, Jacquot, J.P, Krimm, I, Lancelin, J.M. | | Deposit date: | 2004-06-16 | | Release date: | 2005-04-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure and solution NMR dynamics of a D (type II) peroxiredoxin glutaredoxin and thioredoxin dependent: a new insight into the peroxiredoxin oligomerism
Biochemistry, 44, 2005
|
|
1NQ5
 
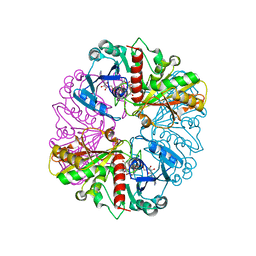 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPT
 
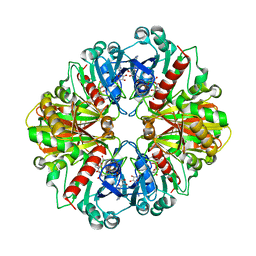 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQA
 
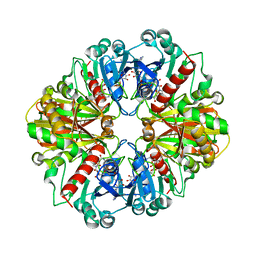 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQO
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-22 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate
Dehydrogenase from Bacillus stearothermophilus with NAD and
D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1EP8
 
 | | CRYSTAL STRUCTURE OF A MUTATED THIOREDOXIN, D30A, FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
1EP7
 
 | | CRYSTAL STRUCTURE OF WT THIOREDOXIN H FROM CHLAMYDOMONAS REINHARDTII | | Descriptor: | THIOREDOXIN CH1, H-TYPE | | Authors: | Menchise, V, Corbier, C, Didierjean, C, Saviano, M, Benedetti, E, Jacquot, J.P, Aubry, A. | | Deposit date: | 2000-03-28 | | Release date: | 2001-12-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism.
Biochem.J., 359, 2001
|
|
