3RZZ
 
 | |
3I58
 
 | |
3I64
 
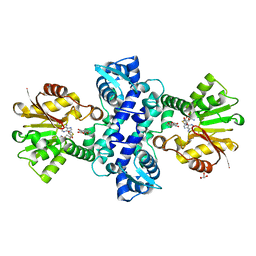 | | Crystal structure of an O-methyltransferase (NcsB1) from neocarzinostatin biosynthesis in complex with S-adenosyl-L-homocysteine (SAH) and 1,4-dihydroxy-2-naphthoic acid (DHN) | | Descriptor: | 1,4-dihydroxy-2-naphthoic acid, GLYCEROL, O-methyltransferase, ... | | Authors: | Cooke, H.A, Bruner, S.D. | | Deposit date: | 2009-07-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis of substrate promiscuity for the SAM-dependent O-methyltransferase NcsB1, involved in the biosynthesis of the enediyne antitumor antibiotic neocarzinostatin.
Biochemistry, 48, 2009
|
|
3I53
 
 | |
3I5U
 
 | |
3KDY
 
 | |
3KDZ
 
 | |
