1Y6U
 
 | |
2KID
 
 | | Solution Structure of the S. Aureus Sortase A-substrate Complex | | Descriptor: | (PHQ)LPA(B27) peptide, CALCIUM ION, Sortase | | Authors: | Suree, N, Liew, C.K, Villareal, V.A, Thieu, W, Fadeev, E.A, Clemens, J.J, Jung, M.E, Clubb, R.T. | | Deposit date: | 2009-05-01 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the Staphylococcus aureus sortase-substrate complex reveals how the universally conserved LPXTG sorting signal is recognized.
J.Biol.Chem., 284, 2009
|
|
2K78
 
 | | Solution Structure of the IsdC NEAT domain bound to Zinc Protoporphyrin | | Descriptor: | Iron-regulated surface determinant protein C, PROTOPORPHYRIN IX CONTAINING ZN | | Authors: | Villareal, V.A, Pilpa, R.M, Robson, S.A, Fadeev, E.A, Clubb, R.T. | | Deposit date: | 2008-08-06 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The IsdC Protein from Staphylococcus aureus Uses a Flexible Binding Pocket to Capture Heme.
J.Biol.Chem., 283, 2008
|
|
2LHR
 
 | |
2OG0
 
 | | Crystal Structure of the Lambda Xis-DNA complex | | Descriptor: | 5'-D(*AP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*AP*TP*AP*C)-3', 5'-D(*GP*TP*AP*TP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*T)-3', Excisionase | | Authors: | Papagiannis, C.V, Sam, M.D, Abbani, M.A, Cascio, D, Yoo, D, Clubb, R.T, Johnson, R.C. | | Deposit date: | 2007-01-04 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fis targets assembly of the xis nucleoprotein filament to promote excisive recombination by phage lambda.
J.Mol.Biol., 367, 2007
|
|
2LN7
 
 | |
2KW8
 
 | |
2EZH
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZK
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, REGULARIZED MEAN STRUCTURE | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2EZI
 
 | | SOLUTION NMR STRUCTURE OF THE IGAMMA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF MU PHAGE TRANSPOSASE, 30 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-07-25 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the I gamma subdomain of the Mu end DNA-binding domain of phage Mu transposase.
J.Mol.Biol., 273, 1997
|
|
2EZL
 
 | | SOLUTION NMR STRUCTURE OF THE IBETA SUBDOMAIN OF THE MU END DNA BINDING DOMAIN OF PHAGE MU TRANSPOSASE, 29 STRUCTURES | | Descriptor: | TRANSPOSASE | | Authors: | Clore, G.M, Clubb, R.T, Schumaker, S, Gronenborn, A.M. | | Deposit date: | 1997-10-04 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Mu end DNA-binding ibeta subdomain of phage Mu transposase: modular DNA recognition by two tethered domains.
EMBO J., 16, 1997
|
|
2H3K
 
 | | Solution Structure of the first NEAT domain of IsdH | | Descriptor: | Haptoglobin-binding surface anchored protein | | Authors: | Pilpa, R.M, Fadeev, E.A, Villareal, V.A, Wong, M.A, Phillips, M, Clubb, R.T. | | Deposit date: | 2006-05-22 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the NEAT (NEAr Transporter) domain from IsdH/HarA: the human hemoglobin receptor in Staphylococcus aureus.
J.Mol.Biol., 360, 2006
|
|
2IEF
 
 | | Structure of the cooperative Excisionase (Xis)-DNA complex reveals a micronucleoprotein filament | | Descriptor: | 15-mer DNA, 19-mer DNA, 34-mer DNA, ... | | Authors: | Abbani, M.A, Papagiannis, C.V, Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2006-09-18 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of the cooperative Xis-DNA complex reveals a micronucleoprotein filament that regulates phage lambda intasome assembly.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1LX8
 
 | | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein | | Descriptor: | Excisionase | | Authors: | Sam, M.D, Papagiannis, C, Connolly, K.M, Corselli, L, Iwahara, J, Lee, J, Phillips, M, Wojciak, J.M, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2002-06-04 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Regulation of directionality in bacteriophage lambda site-specific recombination: structure of the Xis protein
J.Mol.Biol., 324, 2002
|
|
1QPM
 
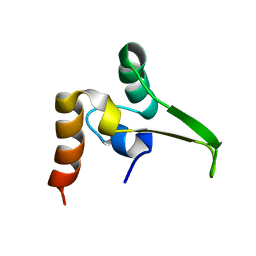 | |
1IJA
 
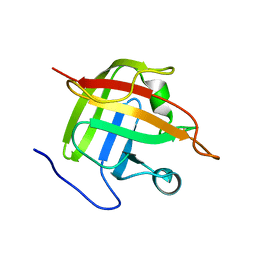 | | Structure of Sortase | | Descriptor: | Sortase | | Authors: | Ilangovan, U, Ton-That, H, Iwahara, J, Schneewind, O, Clubb, R.T. | | Deposit date: | 2001-04-25 | | Release date: | 2001-05-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1KQQ
 
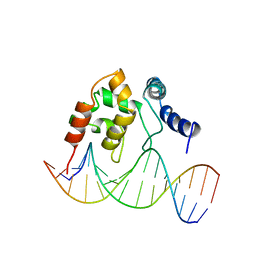 | | Solution Structure of the Dead ringer ARID-DNA Complex | | Descriptor: | 5'-D(*CP*CP*AP*CP*AP*TP*CP*AP*AP*TP*AP*CP*AP*GP*G)-3', 5'-D(*CP*CP*TP*GP*TP*AP*TP*TP*GP*AP*TP*GP*TP*GP*G)-3', DEAD RINGER PROTEIN | | Authors: | Iwahara, J, Iwahara, M, Daughdrill, G.W, Ford, J, Clubb, R.T. | | Deposit date: | 2002-01-07 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA.
EMBO J., 21, 2002
|
|
1KJK
 
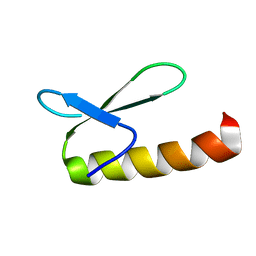 | |
1G4D
 
 | |
1RH6
 
 | | Bacteriophage Lambda Excisionase (Xis)-DNA Complex | | Descriptor: | 5'-D(*CP*TP*AP*TP*GP*TP*AP*GP*TP*CP*TP*GP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*CP*AP*GP*AP*CP*TP*AP*CP*AP*TP*AP*G)-3', Excisionase | | Authors: | Sam, M.D, Cascio, D, Johnson, R.C, Clubb, R.T. | | Deposit date: | 2003-11-13 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the excisionase-DNA complex from bacteriophage lambda.
J.Mol.Biol., 338, 2004
|
|
1TNS
 
 | |
1TNT
 
 | |
