6RP1
 
 | | 1.49 A RESOLUTION OF SPOROSARCINA PASTEURII UREASE INHIBITED IN THE PRESENCE OF NBPTO AT pH 6.5 | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The Impact of pH on Catalytically Critical Protein Conformational Changes: The Case of the Urease, a Nickel Enzyme.
Chemistry, 25, 2019
|
|
6RKG
 
 | | 1.32 A RESOLUTION OF SPOROSARCINA PASTEURII UREASE INHIBITED IN THE PRESENCE OF NBPTO AT pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Benini, S, Ciurli, S. | | Deposit date: | 2019-04-30 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | The Impact of pH on Catalytically Critical Protein Conformational Changes: The Case of the Urease, a Nickel Enzyme.
Chemistry, 25, 2019
|
|
2J8W
 
 | | The crystal structure of cytochrome c' from Rubrivivax gelatinosus at 1.3 A Resolution and pH 8.0 | | Descriptor: | CYTOCHROME C', HEME C | | Authors: | Benini, S, Ciurli, S, Rypniewski, W.R, Wilson, K.S. | | Deposit date: | 2006-10-30 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | High resolution crystal structure of Rubrivivax gelatinosus cytochrome c'.
J. Inorg. Biochem., 102, 2008
|
|
6G48
 
 | | Sporosarcina pasteurii urease inhibited by silver | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-03-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structure of urease inactivated by Ag(i): a new paradigm for enzyme inhibition by heavy metals.
Dalton Trans, 47, 2018
|
|
1IE7
 
 | | PHOSPHATE INHIBITED BACILLUS PASTEURII UREASE CRYSTAL STRUCTURE | | Descriptor: | NICKEL (II) ION, PHOSPHATE ION, UREASE ALPHA SUBUNIT, ... | | Authors: | Benini, S, Rypniewski, W.R, Wilson, K.S, Ciurli, S, Mangani, S. | | Deposit date: | 2001-04-09 | | Release date: | 2001-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based rationalization of urease inhibition by phosphate: novel insights into the enzyme mechanism.
J.Biol.Inorg.Chem., 6, 2001
|
|
5OL4
 
 | | 1.28 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPT | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Phosphoramidothioic O,O-acid, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2017-07-26 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Urease Inhibition in the Presence of N-(n-Butyl)thiophosphoric Triamide, a Suicide Substrate: Structure and Kinetics.
Biochemistry, 56, 2017
|
|
1EAR
 
 | | Crystal structure of Bacillus pasteurii UreE at 1.7 A. Type II crystal form. | | Descriptor: | UREASE ACCESSORY PROTEIN UREE, ZINC ION | | Authors: | Remaut, H, Safarov, N, Ciurli, S, Van Beeumen, J. | | Deposit date: | 2001-07-16 | | Release date: | 2002-01-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone Uree from Bacillus Pasteurii
J.Biol.Chem., 276, 2001
|
|
1EB0
 
 | | Crystal structure of Bacillus pasteurii UreE at 1.85 A, phased by SIRAS. Type I crystal form. | | Descriptor: | UREASE ACCESSORY PROTEIN UREE, ZINC ION | | Authors: | Remaut, H, Safarov, N, Ciurli, S, Van Beeumen, J. | | Deposit date: | 2001-07-17 | | Release date: | 2002-01-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Ni2+ Transport and Assembly of the Urease Active Site by the Metallochaperone Uree from Bacillus Pasteurii
J.Biol.Chem., 276, 2001
|
|
7P6F
 
 | | 1.93 A resolution X-ray crystal structure of the transcriptional regulator SrnR from Streptomyces griseus | | Descriptor: | ACETATE ION, SODIUM ION, Transcriptional regulator SrnR | | Authors: | Mazzei, L, Ciurli, S. | | Deposit date: | 2021-07-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure, dynamics, and function of SrnR, a transcription factor for nickel-dependent gene expression.
Metallomics, 13, 2021
|
|
6HK5
 
 | | X-ray structure of a truncated mutant of the metallochaperone CooJ with a high-affinity nickel-binding site | | Descriptor: | 3,3',3''-phosphoryltripropanoic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Alfano, M, Perard, J, Basset, C, Carpentier, P, Zambelli, B, Timm, J, Crouzy, S, Ciurli, S, Cavazza, C. | | Deposit date: | 2018-09-05 | | Release date: | 2019-03-27 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | The carbon monoxide dehydrogenase accessory protein CooJ is a histidine-rich multidomain dimer containing an unexpected Ni(II)-binding site.
J.Biol.Chem., 294, 2019
|
|
7P7N
 
 | |
7P7O
 
 | | X-RAY CRYSTAL STRUCTURE OF SPOROSARCINA PASTEURII UREASE INHIBITED BY THE GOLD(I)-DIPHOSPHINE COMPOUND Au(PEt3)2Cl DETERMINED AT 1.87 ANGSTROMS | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, OXYGEN ATOM, ... | | Authors: | Mazzei, L, Ciurli, S, Cianci, M, Messori, L, Massai, L. | | Deposit date: | 2021-07-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Medicinal Au(I) compounds targeting urease as prospective antimicrobial agents: unveiling the structural basis for enzyme inhibition.
Dalton Trans, 50, 2021
|
|
5FSE
 
 | | 2.07 A resolution 1,4-Benzoquinone inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Inactivation of Urease by 1,4-Benzoquinone: Chemistry at the Protein Surface.
Dalton Trans, 45, 2016
|
|
5G4H
 
 | | 1.50 A resolution catechol (1,2-dihydroxybenzene) inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, CATECHOL, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-05-13 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Inactivation of Urease by Catechol: Kinetics and Structure.
J.Inorg.Biochem., 166, 2016
|
|
5FSD
 
 | | 1.75 A resolution 2,5-dihydroxybenzensulfonate inhibited Sporosarcina pasteurii urease | | Descriptor: | 1,2-ETHANEDIOL, 2,5-dihydroxybenzenesulfonic acid, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Musiani, F, Ciurli, S. | | Deposit date: | 2016-01-04 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Inactivation of Urease by 1,4-Benzoquinone: Chemistry at the Protein Surface.
Dalton Trans, 45, 2016
|
|
6H8J
 
 | | 1.45 A resolution of Sporosarcina pasteurii urease inhibited in the presence of NBPTO | | Descriptor: | 1,2-ETHANEDIOL, DIAMIDOPHOSPHATE, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-08-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into Urease Inhibition by N-( n-Butyl) Phosphoric Triamide through an Integrated Structural and Kinetic Approach.
J.Agric.Food Chem., 67, 2019
|
|
6I9Y
 
 | | The 2.14 A X-ray crystal structure of Sporosarcina pasteurii urease in complex with Au(I) ions | | Descriptor: | 1,2-ETHANEDIOL, GOLD ION, HYDROXIDE ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-11-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibition Mechanism of Urease by Au(III) Compounds Unveiled by X-ray Diffraction Analysis.
Acs Med.Chem.Lett., 10, 2019
|
|
1K3G
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
1K3H
 
 | | NMR Solution Structure of Oxidized Cytochrome c-553 from Bacillus pasteurii | | Descriptor: | HEME C, cytochrome c-553 | | Authors: | Banci, L, Bertini, I, Ciurli, S, Dikiy, A, Dittmer, J, Rosato, A, Sciara, G, Thompsett, A.R. | | Deposit date: | 2001-10-03 | | Release date: | 2001-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure, backbone mobility, and homology modeling of c-type cytochromes from gram-positive bacteria.
Chembiochem, 3, 2002
|
|
3TJ8
 
 | | Crystal structure of Helicobacter pylori UreE bound to Ni2+ | | Descriptor: | FORMIC ACID, NICKEL (II) ION, Urease accessory protein ureE | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
3TJA
 
 | | Crystal structure of Helicobacter pylori UreE in the apo form | | Descriptor: | CHLORIDE ION, SULFATE ION, Urease accessory protein ureE | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
3TJ9
 
 | | Crystal structure of Helicobacter pylori UreE bound to Zn2+ | | Descriptor: | FORMIC ACID, Urease accessory protein ureE, ZINC ION | | Authors: | Banaszak, K, Bellucci, M, Zambelli, B, Rypniewski, W.R, Ciurli, S. | | Deposit date: | 2011-08-24 | | Release date: | 2011-11-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Crystallographic and X-ray absorption spectroscopic characterization of Helicobacter pylori UreE bound to Ni2+ and Zn2+ reveals a role for the disordered C-terminal arm in metal trafficking.
Biochem.J., 441, 2012
|
|
1JXD
 
 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1JXF
 
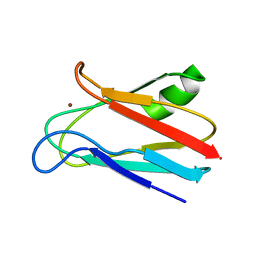 | | SOLUTION STRUCTURE OF REDUCED CU(I) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803 | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Bryant, D.A, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Vila, A.J, Zhao, J. | | Deposit date: | 2001-09-07 | | Release date: | 2001-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of plastocyanin in both oxidation states. Solution structure of the reduced form and comparison with the oxidized state.
J.Biol.Chem., 276, 2001
|
|
1J5D
 
 | | SOLUTION STRUCTURE OF OXIDIZED PARAMAGNETIC CU(II) PLASTOCYANIN FROM SYNECHOCYSTIS PCC6803-MINIMIZED AVERAGE STRUCTURE | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Bertini, I, Ciurli, S, Dikiy, A, Fernandez, C.O, Luchinat, C, Safarov, N, Shumilin, S, Vila, A.J. | | Deposit date: | 2002-04-02 | | Release date: | 2002-04-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The first solution structure of a paramagnetic copper(II) protein: the case of oxidized plastocyanin from the cyanobacterium Synechocystis PCC6803.
J.Am.Chem.Soc., 123, 2001
|
|
